25 February 2023: Original Paper
Detecting Donor-Derived DNA by Real-Time PCR in Recipients Suspected of Graft-Versus-Host-Diseases After Liver Transplantation: A Case Series and Literature Review
Changhee HaDOI: 10.12659/AOT.938287
Ann Transplant 2023; 28:e938287
Abstract
BACKGROUND: Graft-versus-host disease (GVHD) after liver transplantation (LT) is a rare but fatal complication. GVHD diagnosis is usually based on clinical symptoms and pathologic confirmation. However, it is often misdiagnosed due to its non-specific symptoms. Here, we report the detection of donor-cell chimerism using peripheral blood (PB) donor-derived deoxyribonucleic acid (ddDNA) for 3 cases with suspected GVHD after LT (GVHD-LT) through real-time quantitative polymerase chain reaction (qPCR) assay targeting 39 insertions and/or deletions of chromosomes.
MATERIAL AND METHODS: The qPCR assay for detecting donor-cell chimerism was performed for 3 post-LT patients with suspected GVHD using KMRtype® and KMRtrack® assays (GenDx, Netherlands). The mean recipient/donor-cell fraction of informative markers unique to each recipient or donor was calculated.
RESULTS: In Case 1, who received living donor LT (LDLT) from his daughter, initial sign was diarrhea at post-operative day (POD) #23. Case 2 received unrelated deceased donor LT and initial sign was cytopenia at POD #29. Case 3 received LDLT from her son and GVHD associated cytopenia was developed at POD #80. Average PB ddDNA fractions in post-transplant samples of cases 1, 2, and 3 were 39.68%, 78.38%, and 4.76%, respectively. Despite an active treatment including steroid and tumor necrosis factor-α inhibitor, 2 patients (cases 1 and 2) died due to multiple organ failures.
CONCLUSIONS: Early detection of donor-cell chimerism may help halt fatal progression of GVHD-LT. A qPCR test targeting INDEL of chromosomes would be a helpful procedure for timely diagnosis of GVHD.
Keywords: Chimerism, Graft vs Host Disease, Liver Transplantation, Real-Time Polymerase Chain Reaction, Humans, Female, Living Donors, DNA
Background
Graft-versus-host-disease (GVHD) is a rare but fatal complication of liver transplantation (LT). Unlike an incidence of 35–50% after hematopoietic stem-cell transplantation (HSCT), incidence of GVHD after LT (GVHD-LT) is only up to 2%. However, mortality of such cases reaches above 75% [1,2]. GVHD occurs when immunocompetent T cells in the graft recognize host antigens as foreign, derive immune responses, and attack the recipient, leading to extensive tissue damages and life-threatening complications [3]. Reported risk factors for GVHD include human leukocyte antigen (HLA) incompatibility, sex mismatch, and increased recipient age [2–4].
Clinical manifestations of acute GVHD include skin rash, diarrhea, and pancytopenia as organs containing highly proliferating cells such as skin, gastrointestinal tract, and bone marrow (BM) are primary targets of immune reactions [3,5]. These manifestations usually occur at 3 to 5 weeks after LT [1]. Timely diagnosis of GVHD-LT is needed, based on clinical manifestations. However, diagnosis of GVHD-LT faces difficulties because early symptoms are non-specific. Thus, GVHD-LT is often misdiagnosed with other etiologies, such as cytomegalovirus (CMV) infection,
Identification of donor hematopoietic cell chimerism by detecting donor-derived deoxyribonucleic acid (ddDNA) in the recipient’s peripheral blood (PB) is one of the most efficient confirmatory tests of GVHD-LT [1]. It has been an effective method due to its minimal invasiveness and cost-effectiveness [7,8]. Currently, targeting short tandem repeats (STRs) of DNA using polymerase chain reaction (PCR) is the most commonly used method in HSCT settings [9]. Among ddDNA quantification techniques, real-time quantitative polymerase chain reaction (qPCR) using unique single-nucleotide polymorphisms (SNPs) or insertion and/or deletions (INDELs) is a recently introduced method with high sensitivity and laboratory feasibility [3,10]. SNPs are the most abundant form of human genetic variability; they are unique and can be analyzed by qPCR. Despite the advantages of SNPs, these markers have complex assay design [11]. In contrast to SNPs, INDEL-qPCRs have less assay complexity and can better detect target DNA (0.01~0.1%) [10-12] than SNP-qPCR (0.1%) [11,13]. In kidney transplantation (KT) patients, the INDEL-qPCR has high sensitivity and specificity to detect acute rejection [14]. However, there have been no studies using INDEL-qPCR assay in LT or GVHD-LT patients. Thus, the objective of this study was to detect donor-cell chimerism using PB ddDNA through a qPCR assay for 3 cases with suspected of GVHD-LT in a single center based on clinical manifestations and histological findings.
Material and Methods
STUDY SUBJECTS AND IMMUNOSUPPRESSION PROTOCOL:
Our transplantation center performs approximately 150 LT per year, including both living donor LT (LDLT) and deceased donor LT (DDLT). Although GVHD is rare, 3 patients with suspected GVHD were found prospectively between December 2018 and March 2020.
Each patient received basiliximab, an interleukin-2 receptor monoclonal antibody (Simulect, Novartis Pharma AG, Basel, Switzerland) for immunosuppression induction. Post-transplant intravenous (IV) methylprednisolone was administered with gradual dose reduction. It was subsequently changed to oral prednisolone and tapered accordingly.
Tacrolimus and mycophenolate mofetil (MMF) were used for immunosuppression maintenance therapy. MMF was started on the morning of post-operative day (POD) #1. Tacrolimus was begun on the evening of POD #3 with a high dose in the early phase, which was gradually reduced to target trough levels of 8–10 ng/mL at around 1 month after LT. This policy was applied to both LDLT and DDLT.
LABORATORY TESTS:
Laboratory tests, including complete blood count (CBC) and liver function test, were periodically performed after LT, initially 4 times a day. The sampling interval was gradually extended to once-daily during admission. After discharge, patients were checked monthly at routine outpatient department visits.
For infection surveillance, quantitative CMV antigenemia targeting pp65 was tested using direct immunofluorescence method (CINA Kit system, Argene Biosoft, Varilhes, France) 3 times during the first week after transplantation and then weekly during hospitalization. After discharge, patients were monitored monthly during the first year after LT. CMV qPCR testing (Real-Q assay; BioSewoom, Seoul, South Korea) was performed when the patient had leukopenia, according to the clinician’s judgement.
TISSUE BIOPSY:
Simultaneous development of GVHD features led to invasive diagnostic procedures, where trunk punch biopsy was performed for patients with skin rash and colonoscopic biopsy was performed for patients with persistent diarrhea or hematochezia. BM biopsy was not routinely conducted due to its invasiveness despite occurrence of cytopenia. Biopsy specimens were examined after hematoxylin and eosin staining using an Olympus EX51 microscope (Olympus Life Science, Waltham, MA, USA).
REAL-TIME QUANTITATIVE PCR CHIMERISM ASSAY USING POLYMORPHIC INDELS:
PB specimens from the patient and donor were collected into ethylenediaminetetraacetic acid anticoagulant tubes. Genomic DNA was extracted using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). Chimerism analysis was performed using the commercial KMRtype® genotyping kit and KMRtrack® monitoring assays (GenDx, Utrecht, Netherlands) following the manufacturer’s instructions. This analysis consisted of 2 major processes. The first process involved genotyping of pre-transplantation DNA samples with the KMRtype® kit (GenDx). This kit covers 39 non-HLA bi-allelic markers placed along 17 different chromosomes (Table 1). This process enables identification of specific genetic INDEL markers, also referred to as ‘informative markers’, which uniquely exist in either recipient or donor form. The second process was based on detecting chimerism using KMRtrack® monitoring assays (GenDx). Post-transplantation DNA samples were subjected to monitoring of existence of informative markers from the genotyping process. Informative markers in the post-transplantation chimeric mixture were quantified by analyzing amounts of fluorescence of pre- and post-transplantation samples.
Calculations of chimerism fraction were performed using the dedicated software KMRengine® (GenDx). The proportion of informative markers was calculated with a formula based on cycle quantification value (Cq) defined as the number of cycles in which the measured fluorescence reached the threshold. Comparing the Cq value of the marker to the Cq value of the reference assay (ΔCq=Cqmarker-Cqreference) was possible for both pre- and post-transplantation samples. For the reference assay, an invariant housekeeping gene was used. Subsequently, ΔΔCq ((Cqmarker_post-Cqreference_post) - (Cqmarker_pre-Cqreference_pre), the ΔCq between pre- and post-transplantation samples, was calculated. As the Cq value was inversely proportional to the original amount of DNA, the formula using ΔΔCq was obtained and the percentage of the informative marker in the post-transplantation sample was 100/2ΔΔCq. A schematic diagram of the qPCR chimerism assay and results of 1 representative case are described in Figure 1.
LITERATURE REVIEW:
We searched the literatures on GVHD-LT in PubMed, KoreaMed, and Google Scholar databases reported between January 2002 and December 2021. Reports were filtered using keywords such as LT, GVHD, and chimerism. Duplicates were removed and 66 articles written in either English or Korean were initially selected. Among them, studies that clearly stated the tested specimen (PB or BM), detection method, test day, and chimerism levels were carefully selected.
Results
CASE 1:
A 66-year-old man with underlying diabetes mellitus (DM) received LDLT in December 2018 due to liver cirrhosis (LC) and hepatocellular carcinoma (HCC). The donor was a 38-year-old daughter of a recipient with a homozygous HLA type in HLA-A, HLA-B, HLA-C, HLA-DQ, and HLA-DR loci, matching the haplotype of the recipient. Complement-dependent cytotoxicity (CDC) cross-match results were negative in the pre-operative work-up. On POD #21, the patient developed CMV antigenemia (7/200 000 white blood cells [WBCs]). According to our institution’s protocol starting preemptive therapy (IV ganciclovir) for cases with >5/200 000 WBCs, IV ganciclovir treatment was started. CMV qPCR was not tested in this patient based on the clinician’s decision. The patient presented with diarrhea and skin rash on POD #23 and POD #24, respectively. Antibiotics ampicillin/sulbactam with cefotaxime were started owing to mild fever. When GVHD was suspected with the development of neutropenia (WBC: 670/μL [reference value: 3800–10 580/uL], absolute neutrophil count (ANC): 520/μL [1570–8300/uL]), persistent diarrhea, and expanding rash, a skin biopsy was performed on POD #29 (Figure 3). Ganciclovir was changed to foscarnet and antibiotics were escalated to piperacillin-tazobactam owing to neutropenic fever. Cessation of immunosuppressants was implemented followed by a mini-steroid pulse (IV methylprednisolone 500 mg and daily half-tapering) with 25 mg of etanercept, a TNF-α inhibitor. On POD #30, the PB of the recipient was acquired to perform the qPCR assay. In this assay, 2 informative markers (both specific to the recipient) were selected and the mean fraction of PB ddDNA in recipient DNA was found to be 39.68% (Table 2). However, the patient developed sepsis and multi-organ failure and died on POD #32.
CASE 2:
A 49-year-old man with DM received liver transplantation from an unrelated deceased donor in October 2019 owing to alcoholic LC. He had combined DM nephropathy and hepatorenal syndrome with a Model-for-End-Stage Liver Disease score of 36. The pre-operative work-up showed a 24-year-old male donor having a heterozygous HLA type in HLA-A, HLA-B, and homozygous HLA-DR loci with 2 mismatched alleles in HLA-B and HLA-DR. The CDC cross-match result was negative. After transplantation, the patient recovered. He was discharged on POD #22 with scheduled hemodialysis due to sustained kidney dysfunction. On POD #29, the patient visited the emergency room (ER) because of sustained fever. He was admitted to the isolated area of the intensive care unit (ICU) due to positivity of influenza type B antigen. At this time, liver and chest CT showed no specific fever focus. However, CBC showed pancytopenia (WBC: 290/μL [3800–10 580/uL], ANC: 20/μL [1570–8300/uL], hemoglobin (Hb): 7.9 g/dL [13.6–17.4 g/dL], and platelet: 64 000/μL [141 000–316 000/uL]). CMV qPCR was performed on POD #35 and #39, and showed positive results (9340 IU/mL and 5115 IU/mL, respectively). With the development of diarrhea and rash on the whole body, a skin biopsy was performed on POD #37 (Figure 3). With suspicion of GVHD, immunosuppressive agents were reduced and 200 mg hydrocortisone was injected followed by twice-a-day 32 mg IV methylprednisolone. Etanercept was also administered at 25 mg twice a week. On the next day of skin biopsy, the patient’s PB was sampled to perform the qPCR assay. Two informative markers (1 specific to the recipient and 1 to the donor) were selected and the mean fraction of PB ddDNA in recipient was 78.38%. Unfortunately, the patient died due to septic shock on POD #41.
CASE 3:
A 69-year-old woman with underlying hepatitis B virus infection received LDLT in November 2019 due to LC and HCC. The donor was her son aged 43 years with a heterozygous HLA type in HLA-A, HLA-B, and HLA-DR loci having 2 mismatched alleles, 1 each in HLA-A and HLA-B. The patient’s panel reactive antibody screening was positive for both classes I and II without the presence of donor-specific antibodies. The patient was discharged on POD #23 without post-operative complications. In CMV surveillance in the outpatient clinic, she developed CMV antigenemia (32/200 000 WBCs) and was re-admitted to treat CMV reactivation on POD #33. After diagnosis of CMV gastritis by endoscopic evaluation, she received IV ganciclovir for 16 days, which was changed to valganciclovir (POD #49). Oral valganciclovir was continued for 35 days (until POD #83) based on 8 weeks of antiviral treatment protocol. This patient showed mild pancytopenia (POD #34, WBC 2540/uL [3150–8630/uL], ANC 2160/uL [1570–8300/uL], Hb 11.1 g/dL [11.2–14.8g/dL], platelet 105 000/uL [138 000–347 000/μL]) during the initial phase (day 2) of ganciclovir administration, but her leukopenia and thrombocytopenia were recovered (POD #64 and #65, respectively). On POD #80, the patient visited the ER because of sudden development of nausea, vomiting, and a heart-burn sensation. Laboratory results showed a moderate pancytopenia (WBC: 1630/μL [3150–8630/uL], ANC: 1210/μL [1570–8300/uL], Hb: 9.1 g/dL [11.2–14.8g/dL], and platelet: 19 000/μL [138 000–347 000/μL]). CMV antigenemia showed negative result and CMV-associated gastritis finding was not observed in EGD. On the next day, the patient developed hematochezia and diarrhea, followed by suspected GVHD at sigmoidoscopy (Figure 2). After confirmation of GVHD, a moderate-dose steroid was administered with cessation of the immunosuppressive agent. Methylprednisolone 60 mg was administered twice a day for 1 week followed by tapering based on the patient’s symptoms and laboratory test results. On POD #90, the PB of the recipient was sampled to perform the qPCR assay. Seven informative markers (4 specific to the recipient and 3 to the donor) were selected. The mean fraction of PB ddDNA in the recipient was 4.76%. Within 2 weeks, the patient’s laboratory findings and symptoms recovered gradually. Currently, the patient is under routine follow-up without any specific problems.
LITERATURE REVIEW OF CHIMERISM STUDIES IN GVHD AFTER LT:
Among 66 previous reports, 12 studies were selected and summarized along with our study in Table 3. The number of patients in each study ranged from 1 to 3, with both living and deceased donor type LT. Our study was the only study using the INDEL-qPCR method, while PCR analysis using STR markers (STR-PCR) was the most commonly utilized chimerism detection method. Chimerism level showed a wide range of 3% to 96%. Almost all previous reports showed chimerism level of above 5%, the detection sensitivity of STR [9]. Chimerism test days were also variable, ranging from POD #7 to POD #240. Most cases were confirmed in the first 2 months after surgery. Among 17 patients, including our study, 10 (58.8%) were dead.
Discussion
GVHD after solid organ transplantation is still a fatal complication having an abrupt onset. Other differential diagnoses, including CMV infection and adverse drug reactions, interfere with early diagnosis and treatment of GVHD [1,2,6,15], and in our study all 3 patients experienced CMV antigenemia. In the case of an antiviral agent, drug-induced cytopenia has a high occurrence rate, making GVHD diagnosis more demanding. One study showed ganciclovir-induced leukopenia occurrence in 28.6% of the patients at a mean of 12.8 days after initiation of ganciclovir [16], and another showed most drug-induced adverse events occur within 30 days [17]. In LT cases, leukopenia related to ganciclovir/valganciclovir showed an incidence of 45.3% [18]. In addition to the challenges in GVHD diagnosis, effective treatment recommendations of GVHD-LT are currently unavailable and the mortality rate remains high [1]. After the discovery of TNF-α inhibitor, which activates regulatory T cells and suppresses GVHD progression [19], recent studies have used TNF-α inhibitor combined with steroid for GVHD-LT patients and shown better survival rates [6,20]. Another case report has shown that using Ruxolitinib, a JAK I/II inhibitor, can improve survival of GVHD-LT patients who have steroid and TNF-α inhibitor resistance [21], but these results were obtained from a small number of cases in heterogeneous patients [1,6]. A standard treatment regimen for GVHD-LT has not been established yet. Owing to limited treatment options, early diagnosis of GVHD along with early immunomodulation, ICU monitoring, supportive care, and infection control is crucial to reduce mortality [22,23]. Predicting GVHD occurrence by estimating risk factors by monitoring laboratory findings and clinical manifestations is also needed.
Risk factors of acute GVHD (HLA incompatibility, female donor for male recipient, old age [2–4]), and reported risk factors of GVHD-LT such as recipient age (>50 years), donor–recipient age difference (>20 years), younger donor age, HCC, and glucose intolerance should also be carefully monitored [1,24,25]. Reviewing our 3 recipient–donor pairs, 2 recipients were elderly (cases 1 and 3), 2 had DM (cases 1 and 2), and all 3 pairs had major age differences. In a Japanese GVHD-LT study, donor-dominant one-way HLA matching, implying a considerable risk of GVHD, showed frequent fatal outcomes [26]. Of note, the donor in case 1 was homozygous for HLA-A, HLA-B, HLA-C, HLA-DQ, and HLA-DR haplotypes of the recipient, implying that the donor’s immune system did not recognize the recipient’s different haplotype as non-self and entailed potential risk for GVHD. Among our cases, even with active intervention including cessation of immunosuppressant and administration of etanercept, patients 1 and 2 eventually died within 2 days after a shock-state. Both had presented with more severe signs and rapid progression. Due to disturbing factors such as ganciclovir use at the presentation of neutropenia (case 1) and influenza positivity along with fever (case 2), suspicion and treatment were delayed. The patient who survived (case 3) after increasing the steroid dose had suffered from relatively mild manifestations of GVHD signs and symptoms along with low ddDNA level (4.76%, INDEL-qPCR method), which might have led to a better prognosis. This patient had experienced pancytopenia 2 times. While an initial pancytopenia at POD #34 might be related with CMV infection rather than ganciclovir treatment, the second pancytopenia, which occurred at POD #80, was likely a GVHD manifestation, not an adverse effect of antiviral treatment.
An efficient laboratory test to detect ddDNA is needed for early diagnosis to prevent multi-organ involvement of GVHD. To date, several methods have been developed. Fluorescent in situ hybridization analysis with X/Y chromosome-specific probe can detect, quantify, and locate chimerism of small minor cell populations using peripheral blood or biopsy samples [27]. Flow cytometry analysis using anti-HLA-antibodies is also highly sensitive for monitoring chimerism, enabling sub-population analysis by distinguishing major histocompatibility complex antigens in an HLA-mismatched transplantation setting [28,29]. Currently, STR-PCR is the most common assay used for detecting chimerism [9] because it is highly informative and had good quantification accuracy and reproducibility [30,31]. As 2–6 nucleotide tandem repeats are highly polymorphic and distributed along the whole genome, STR markers can well differentiate between recipients and donors, so the STR method is currently in wide use for monitoring HSCT patients. It can also be applied to LT patients. However, the limitation of STR-PCR is its relatively low sensitivity, being approximately 5% [9,30,32]. In patients with ddDNA greater than 5%, STR-PCR is sensitive enough with sufficient accuracy, while STR-PCR may show negative results when ddDNA is less than 5% owing to PCR competition and plateau biases [9,30,33]. Another shortcoming of STR-PCR is that it is a labor-intensive and time-consuming procedure [31,32,34], making early detection of GVHD challenging.
After reviewing previous chimerism studies of GVHD-LT, STR-PCR was found to be the most commonly used method (Table 3), with almost all reports showing chimerism level above the detection sensitivity of STR-PCR [9]. Among patients with clinical progression in previous reports, surviving patients showed relatively low levels of chimerism or high levels of chimerism detected at a relatively late period (chimerism level 34%, detected at POD #48) [21]. To the best of our knowledge, our case series is the first report of GVHD-LT proven by INDEL-qPCR assay, a relatively powerful and new method compared to STR-PCR. This assay targets multiple SNPs [3,11] or INDEL polymorphisms (INDEL-qPCR) [10], which are either present or absent in the individual genotype. These methods are capable of rapid detection of target DNA, along with higher sensitivity (0.01~0.1%) and accuracy [10,30–33]. In addition, qPCR has a shorter data analysis time compared to STR-PCR [32]. It also has a rapid laboratory procedure time (qPCR: 3–4 hours [30,34] vs STR-PCR: ~2 days [31,34]). Among the 2 targets (multiple SNPs and INDEL polymorphisms), INDEL markers were more broadly used due to their simplicity [12].
Since most patients with GVHD-LT have leukopenia, the amount of DNA extraction might affect assay performance. Some studies have suggested that INDEL-qPCR requires higher amounts of input DNA than STR-PCR does [31,32]. However, another study has shown that PCR efficiency does not change even after a 4-fold reduction in input DNA [35]. In our study, we were able to detect low chimerism levels (below 5%) using INDEL-qPCR, even though the patients had pancytopenia. Another advantage of ddDNA is its usefulness as a less-invasive GVHD-specific monitoring marker. Although pathologic diagnosis is confirmatory to diagnose GVHD, frequent testing could not be performed due to its invasiveness. Compared to BM, skin, and colonoscopic biopsies, venous blood sampling is much less invasive, especially considering a patient’s cytopenic state, infection risk, and immunized state. In addition, venous sampling routes have been already prepared in these patients, thereby having an advantage of frequent monitoring. Since routine performance of invasive tests has many obstacles, it should be considered cautiously. Blood-based non-invasive ddDNA testing using INDEL-qPCR assay is a better and more timely monitoring option for GVHD-LT.
Since our study has limitations such as a small number of case series, multicenter evaluation and identification of more GVHD-LT cases diagnosed by INDEL-qPCR chimerism assay will allow better understanding and clues about the disease. In future clinical practice, adapting INDEL-qPCR chimerism assay could also be considered for sequential monitoring after diagnosis and is for making a decision to initiate GVHD-LT treatment.
Conclusions
In this study, we presented 3 cases of GVHD-LT confirmed by INDEL-qPCR chimerism assay, and a literature review. Since early detection of low-level ddDNA chimerism can provide critical clues for the development and management of GVHD-LT, detailed monitoring of patients’ clinical manifestations and utilizing INDEL-qPCR as a non-invasive, rapid laboratory test would be beneficial.
Figures
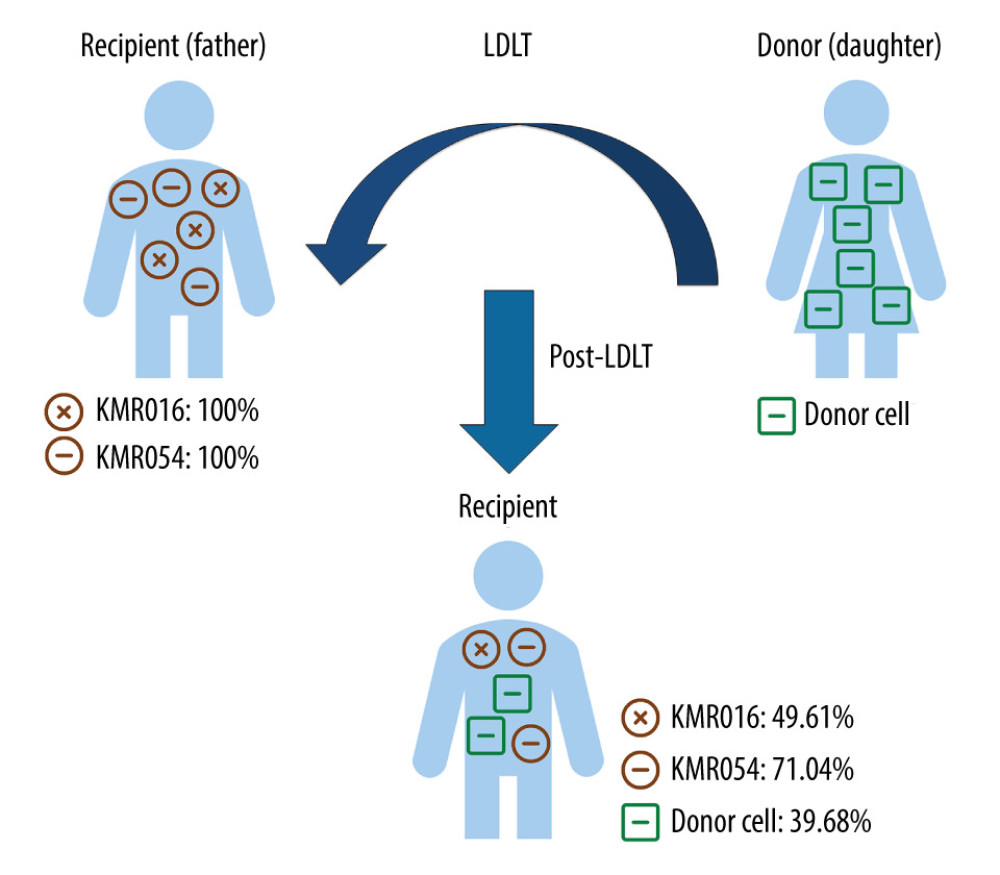 Figure 1. Chimerism assay results for pre- and post-transplantation samples using informative markers of recipient in Case 1 (KMR016 and KMR054). Figure created using PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). LDLT – living donor liver transplantation.
Figure 1. Chimerism assay results for pre- and post-transplantation samples using informative markers of recipient in Case 1 (KMR016 and KMR054). Figure created using PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). LDLT – living donor liver transplantation. 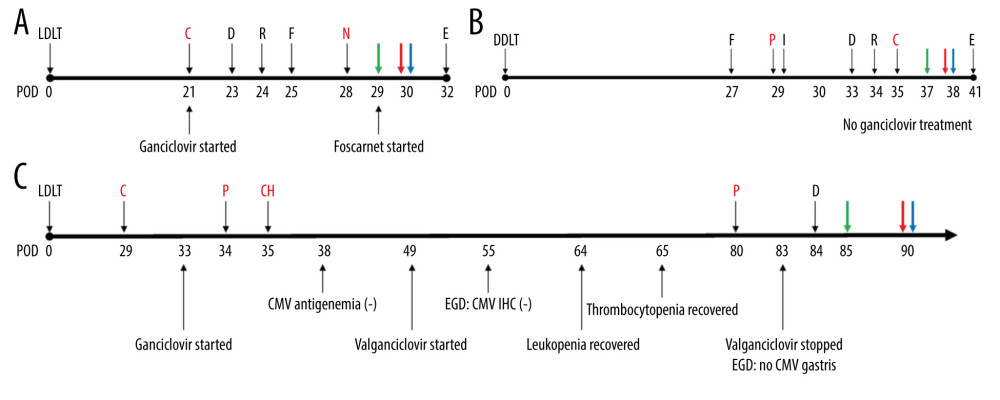 Figure 2. Timeline of recipients’ medical history and clinical courses (post-operative date unscaled); (A) Case 1, (B) Case 2, and (C) Case 3. Green arrow – skin or sigmoidoscopic biopsy; Red arrow – initiation of graft-versus-host disease treatment; Blue arrow – chimerism assay. Cytopenia and CMV related events are in red font. Figure created using PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). CMV – cytomegalovirus; POD – post-operative date; LDLT – living donor liver transplantation; DDLT – deceased donor liver transplantation; C – CMV antigenemia or qPCR (+); D – diarrhea; R – rash; F – fever; N – neutropenia; E – died; P – pancytopenia; I – Influenza positivity; IHC – immunohistochemistry; CH – CMV IHC (+); EGD – esophagogastroduodenoscopy; qPCR – real-time quantitative polymerase chain reaction.
Figure 2. Timeline of recipients’ medical history and clinical courses (post-operative date unscaled); (A) Case 1, (B) Case 2, and (C) Case 3. Green arrow – skin or sigmoidoscopic biopsy; Red arrow – initiation of graft-versus-host disease treatment; Blue arrow – chimerism assay. Cytopenia and CMV related events are in red font. Figure created using PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). CMV – cytomegalovirus; POD – post-operative date; LDLT – living donor liver transplantation; DDLT – deceased donor liver transplantation; C – CMV antigenemia or qPCR (+); D – diarrhea; R – rash; F – fever; N – neutropenia; E – died; P – pancytopenia; I – Influenza positivity; IHC – immunohistochemistry; CH – CMV IHC (+); EGD – esophagogastroduodenoscopy; qPCR – real-time quantitative polymerase chain reaction. 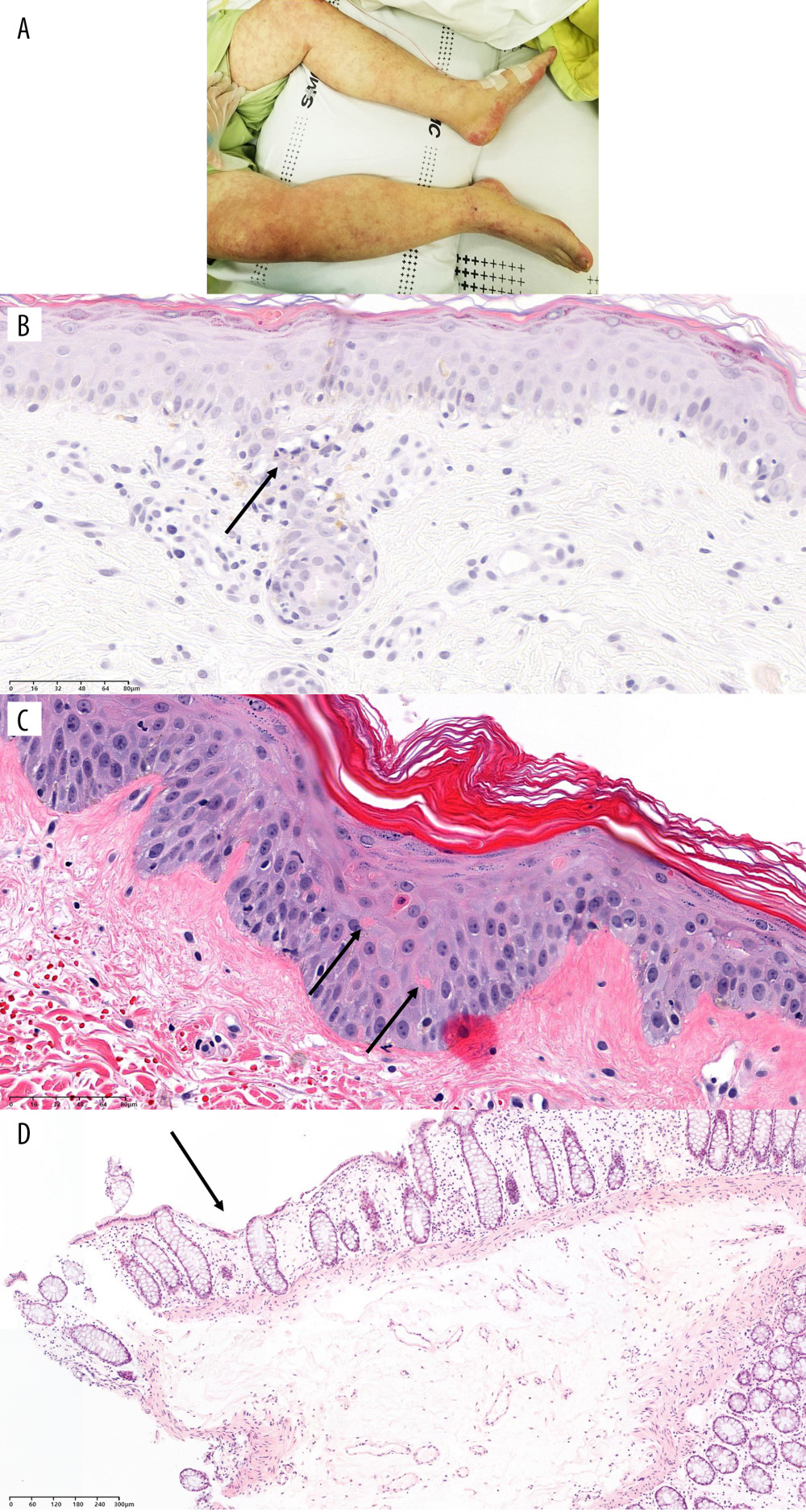 Figure 3. Physical and pathological findings of patients suspected to have graft-versus-host diseases and confirmation of donor chimerism. (A) Skin rash of case 1. (B) Trunk tissue biopsy findings of case 1 showing vacuolization (arrow) and basal cell degeneration. (C) Trunk tissue biopsy findings of case 2 showing an eosinophilic body at the lower layer of the epidermis (arrows). (D) Colonoscopic biopsy findings of case 3 exhibiting ulcer (arrow) and decreased number of crypts with size variation. B–D: hematoxylin and eosin staining. Scale bar is located in the lower-left corner.
Figure 3. Physical and pathological findings of patients suspected to have graft-versus-host diseases and confirmation of donor chimerism. (A) Skin rash of case 1. (B) Trunk tissue biopsy findings of case 1 showing vacuolization (arrow) and basal cell degeneration. (C) Trunk tissue biopsy findings of case 2 showing an eosinophilic body at the lower layer of the epidermis (arrows). (D) Colonoscopic biopsy findings of case 3 exhibiting ulcer (arrow) and decreased number of crypts with size variation. B–D: hematoxylin and eosin staining. Scale bar is located in the lower-left corner. References
1. Murali AR, Chandra S, Stewart Z, Graft versus host disease after liver transplantation in adults: A case series, review of literature, and an approach to management: Transplantation, 2016; 100(12); 2661-70
2. Elsiesy H, Ibrahim A, Selim K, Graft-versus-host disease after liver transplantation: A single-center case series: Ann Transplant, 2015; 20; 397-401
3. Zeiser R, Blazar BR, Acute graft-versus-host disease – biologic process, prevention, and therapy: N Engl J Med, 2017; 377(22); 2167-79
4. Flowers ME, Inamoto Y, Carpenter PA, Comparative analysis of risk factors for acute graft-versus-host disease and for chronic graft-versus-host disease according to National Institutes of Health consensus criteria: Blood, 2011; 117(11); 3214-19
5. Vadakekolathu J, Rutella S, T-cell manipulation strategies to prevent graft-versus-host disease in haploidentical stem cell transplantation: Biomedicines, 2017; 5(2); 33
6. Thin L, Macquillan G, Adams L, Acute graft-versus-host disease after liver transplant: novel use of etanercept and the role of tumor necrosis factor alpha inhibitors: Liver Transpl, 2009; 15(4); 421-26
7. Gielis EM, Ledeganck KJ, De Winter BY, Cell-free DNA: An upcoming biomarker in transplantation: Am J Transplant, 2015; 15(10); 2541-51
8. McClure T, Goh SK, Cox D, Donor-specific cell-free DNA as a biomarker in liver transplantation: A review: World J Transplant, 2020; 10(11); 307-19
9. Clark JR, Scott SD, Jack AL, Monitoring of chimerism following allogeneic haematopoietic stem cell transplantation (HSCT): Technical recommendations for the use of short tandem repeat (STR) based techniques, on behalf of the United Kingdom National External Quality Assessment Service for Leucocyte Immunophenotyping Chimerism Working Group: Br J Haematol, 2015; 168(1); 26-37
10. Kim SY, Jeong MH, Park N, Chimerism monitoring after allogeneic hematopoietic stem cell transplantation using quantitative real-time PCR of biallelic insertion/deletion polymorphisms: J Mol Diagn, 2014; 16(6); 679-88
11. Gineikiene E, Stoskus M, Griskevicius L, Single nucleotide polymorphism-based system improves the applicability of quantitative PCR for chimerism monitoring: J Mol Diagn, 2009; 11(1); 66-74
12. Jiménez-Velasco A, Barrios M, Román-Gómez J, Reliable quantification of hematopoietic chimerism after allogeneic transplantation for acute leukemia using amplification by real-time PCR of null alleles and insertion/deletion polymorphisms: Leukemia, 2005; 19(3); 336-43
13. Eshel R, Vainas O, Shpringer M, Naparstek E, Highly sensitive patient-specific real-time PCR SNP assay for chimerism monitoring after allogeneic stem cell transplantation: Lab Hematol, 2006; 12(1); 39-46
14. Dauber EM, Kollmann D, Kozakowski N, Quantitative PCR of INDELs to measure donor-derived cell-free DNA-a potential method to detect acute rejection in kidney transplantation: a pilot study: Transpl Int, 2020; 33(3); 298-309
15. Perri R, Assi M, Talwalkar J, Graft vs. host disease after liver transplantation: A new approach is needed: Liver Transpl, 2007; 13(8); 1092-99
16. Matsumoto K, Shigemi A, Ikawa K, Risk factors for ganciclovir-induced thrombocytopenia and leukopenia: Biol Pharm Bull, 2015; 38(2); 235-38
17. Ando G, Taguchi K, Enoki Y, Evaluation of the expression time of ganciclovir-induced adverse events using JADER and FAERS: Biol Pharm Bull, 2019; 42(11); 1799-804
18. Wiltshire H, Paya CV, Pescovitz MD, Pharmacodynamics of oral ganciclovir and valganciclovir in solid organ transplant recipients: Transplantation, 2005; 79(11); 1477-83
19. Mancusi A, Piccinelli S, Velardi A, Pierini A, The effect of TNF-α on regulatory T cell function in graft-versus-host disease: Front Immunol, 2018; 9; 356
20. Piton G, Larosa F, Minello A, Infliximab treatment for steroid-refractory acute graft-versus-host disease after orthotopic liver transplantation: A case report: Liver Transpl, 2009; 15(7); 682-85
21. Endo Y, Oshima G, Hibi T, Achievement of durable and complete remission of graft-versus-host disease after liver transplantation with ruxolitinib: A case report: Transplantation, 2019; 103(11); e375-e77
22. Kang WH, Hwang S, Song GW, Acute graft-vs-host disease after liver transplantation: Experience at a high-volume liver transplantation center in Korea: Transplant Proc, 2016; 48(10); 3368-72
23. Rogulj IM, Deeg J, Lee SJ, Acute graft versus host disease after orthotopic liver transplantation: J Hematol Oncol, 2012; 5; 50
24. Elfeki MA, Pungpapong S, Genco PV, Graft-versus-host disease after orthotopic liver transplantation: multivariate analysis of risk factors: Clin Transplant, 2015; 29(12); 1063-66
25. Wood A, Eghtesad B, Lindenmeyer CC, Graft-versus-host disease after liver transplantation: Clin Liver Dis (Hoboken), 2020; 15(2); 81-84
26. Kamei H, Oike F, Fujimoto Y, Fatal graft-versus-host disease after living donor liver transplantation: Differential impact of donor-dominant one-way HLA matching: Liver Transpl, 2006; 12(1); 140-45
27. Meves A, el-Azhary RA, Talwalkar JA, Acute graft-versus-host disease after liver transplantation diagnosed by fluorescent in situ hybridization testing of skin biopsy specimens: J Am Acad Dermatol, 2006; 55(4); 642-46
28. Taylor AL, Gibbs P, Sudhindran S, Monitoring systemic donor lymphocyte macrochimerism to aid the diagnosis of graft-versus-host disease after liver transplantation: Transplantation, 2004; 77(3); 441-46
29. Schumm M, Feuchtinger T, Pfeiffer M, Flow cytometry with anti HLA-antibodies: A simple but highly sensitive method for monitoring chimerism and minimal residual disease after HLA-mismatched stem cell transplantation: Bone Marrow Transplant, 2007; 39(12); 767-73
30. Tyler J, Kumer L, Fisher C, Personalized chimerism test that uses selection of short tandem repeat or quantitative PCR depending on patient’s chimerism status: J Mol Diagn, 2019; 21(3); 483-90
31. Alizadeh M, Bernard M, Danic B, Quantitative assessment of hematopoietic chimerism after bone marrow transplantation by real-time quantitative polymerase chain reaction: Blood, 2002; 99(12); 4618-25
32. Frankfurt O, Zitzner JR, Tambur AR, Real-time qPCR for chimerism assessment in allogeneic hematopoietic stem cell transplants from unrelated adult and double umbilical cord blood: Hum Immunol, 2015; 76(2–3); 155-60
33. Ahci M, Stempelmann K, Buttkereit U, Clinical utility of quantitative PCR for chimerism and engraftment monitoring after allogeneic stem cell transplantation for hematologic malignancies: Biol Blood Marrow Transplant, 2017; 23(10); 1658-68
34. Navarro-Bailón A, Carbonell D, Escudero A, Short tandem repeats (STRs) as biomarkers for the quantitative follow-up of chimerism after stem cell transplantation: Methodological considerations and clinical application: Genes (Basel), 2020; 11(9); 993
35. Bach C, Tomova E, Goldmann K, Monitoring of hematopoietic chimerism by real-time quantitative PCR of micro insertions/deletions in samples with low DNA quantities: Transfus Med Hemother, 2015; 42(1); 38-45
36. Wang L, Yang B, Wei L, Acute graft-versus-host disease after liver transplantation in a close contact with COVID-19: A case report: Transpl Immunol, 2021; 68; 101435
37. Kim KJ, Lee TB, Yang KH, Temporary cessation of immunosuppression for infection may contribute to the development of graft-vs-host disease after ABO-incompatible living donor liver transplantation: A case report: Transplant Proc, 2019; 51(9); 3136-39
38. Chaib E, Silva FD, Figueira ER, Graft-versus-host disease after liver transplantation: Clinics (Sao Paulo), 2011; 66(6); 1115-18
39. Shimizu T, Hayashi M, Inoue Y, Acute graft-versus-host disease after living donor liver transplantation with donor-dominant one-way human leukocyte antigen matching at two Loci: Transplantation, 2010; 89(9); 1164-66
40. Kanehira K, Riegert-Johnson DL, FISH diagnosis of acute graft-versus-host disease following living-related liver transplant: J Mol Diagn, 2009; 11(4); 355-58
41. Cho EH, Suh KS, Yang SH, Acute graft versus host disease following living donor liver transplantation: first Korean report: Hepatogastroenterology, 2007; 54(79); 2120-22
42. Chinnakotla S, Smith DM, Domiati-Saad R, Acute graft-versus-host disease after liver transplantation: Role of withdrawal of immunosuppression in therapeutic management: Liver Transpl, 2007; 13(1); 157-61
43. Pollack MS, Speeg KV, Callander NS, Severe, late-onset graft-versus-host disease in a liver transplant recipient documented by chimerism analysis: Hum Immunol, 2005; 66(1); 28-31
Figures
 Figure 1. Chimerism assay results for pre- and post-transplantation samples using informative markers of recipient in Case 1 (KMR016 and KMR054). Figure created using PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). LDLT – living donor liver transplantation.
Figure 1. Chimerism assay results for pre- and post-transplantation samples using informative markers of recipient in Case 1 (KMR016 and KMR054). Figure created using PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). LDLT – living donor liver transplantation. Figure 2. Timeline of recipients’ medical history and clinical courses (post-operative date unscaled); (A) Case 1, (B) Case 2, and (C) Case 3. Green arrow – skin or sigmoidoscopic biopsy; Red arrow – initiation of graft-versus-host disease treatment; Blue arrow – chimerism assay. Cytopenia and CMV related events are in red font. Figure created using PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). CMV – cytomegalovirus; POD – post-operative date; LDLT – living donor liver transplantation; DDLT – deceased donor liver transplantation; C – CMV antigenemia or qPCR (+); D – diarrhea; R – rash; F – fever; N – neutropenia; E – died; P – pancytopenia; I – Influenza positivity; IHC – immunohistochemistry; CH – CMV IHC (+); EGD – esophagogastroduodenoscopy; qPCR – real-time quantitative polymerase chain reaction.
Figure 2. Timeline of recipients’ medical history and clinical courses (post-operative date unscaled); (A) Case 1, (B) Case 2, and (C) Case 3. Green arrow – skin or sigmoidoscopic biopsy; Red arrow – initiation of graft-versus-host disease treatment; Blue arrow – chimerism assay. Cytopenia and CMV related events are in red font. Figure created using PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). CMV – cytomegalovirus; POD – post-operative date; LDLT – living donor liver transplantation; DDLT – deceased donor liver transplantation; C – CMV antigenemia or qPCR (+); D – diarrhea; R – rash; F – fever; N – neutropenia; E – died; P – pancytopenia; I – Influenza positivity; IHC – immunohistochemistry; CH – CMV IHC (+); EGD – esophagogastroduodenoscopy; qPCR – real-time quantitative polymerase chain reaction. Figure 3. Physical and pathological findings of patients suspected to have graft-versus-host diseases and confirmation of donor chimerism. (A) Skin rash of case 1. (B) Trunk tissue biopsy findings of case 1 showing vacuolization (arrow) and basal cell degeneration. (C) Trunk tissue biopsy findings of case 2 showing an eosinophilic body at the lower layer of the epidermis (arrows). (D) Colonoscopic biopsy findings of case 3 exhibiting ulcer (arrow) and decreased number of crypts with size variation. B–D: hematoxylin and eosin staining. Scale bar is located in the lower-left corner.
Figure 3. Physical and pathological findings of patients suspected to have graft-versus-host diseases and confirmation of donor chimerism. (A) Skin rash of case 1. (B) Trunk tissue biopsy findings of case 1 showing vacuolization (arrow) and basal cell degeneration. (C) Trunk tissue biopsy findings of case 2 showing an eosinophilic body at the lower layer of the epidermis (arrows). (D) Colonoscopic biopsy findings of case 3 exhibiting ulcer (arrow) and decreased number of crypts with size variation. B–D: hematoxylin and eosin staining. Scale bar is located in the lower-left corner. Tables
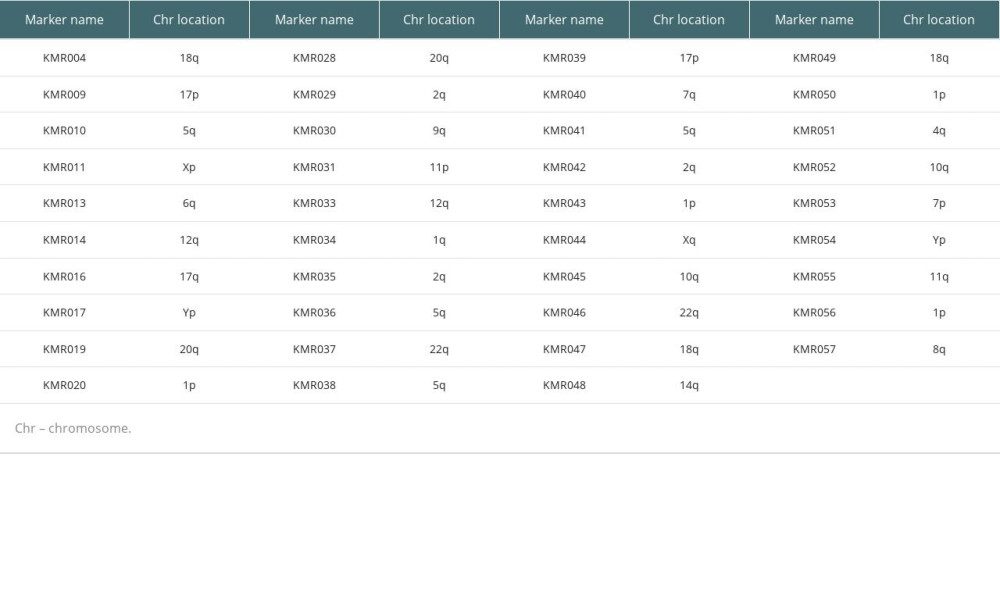 Table 1. List of 39 informative markers and their chromosomal locations.
Table 1. List of 39 informative markers and their chromosomal locations.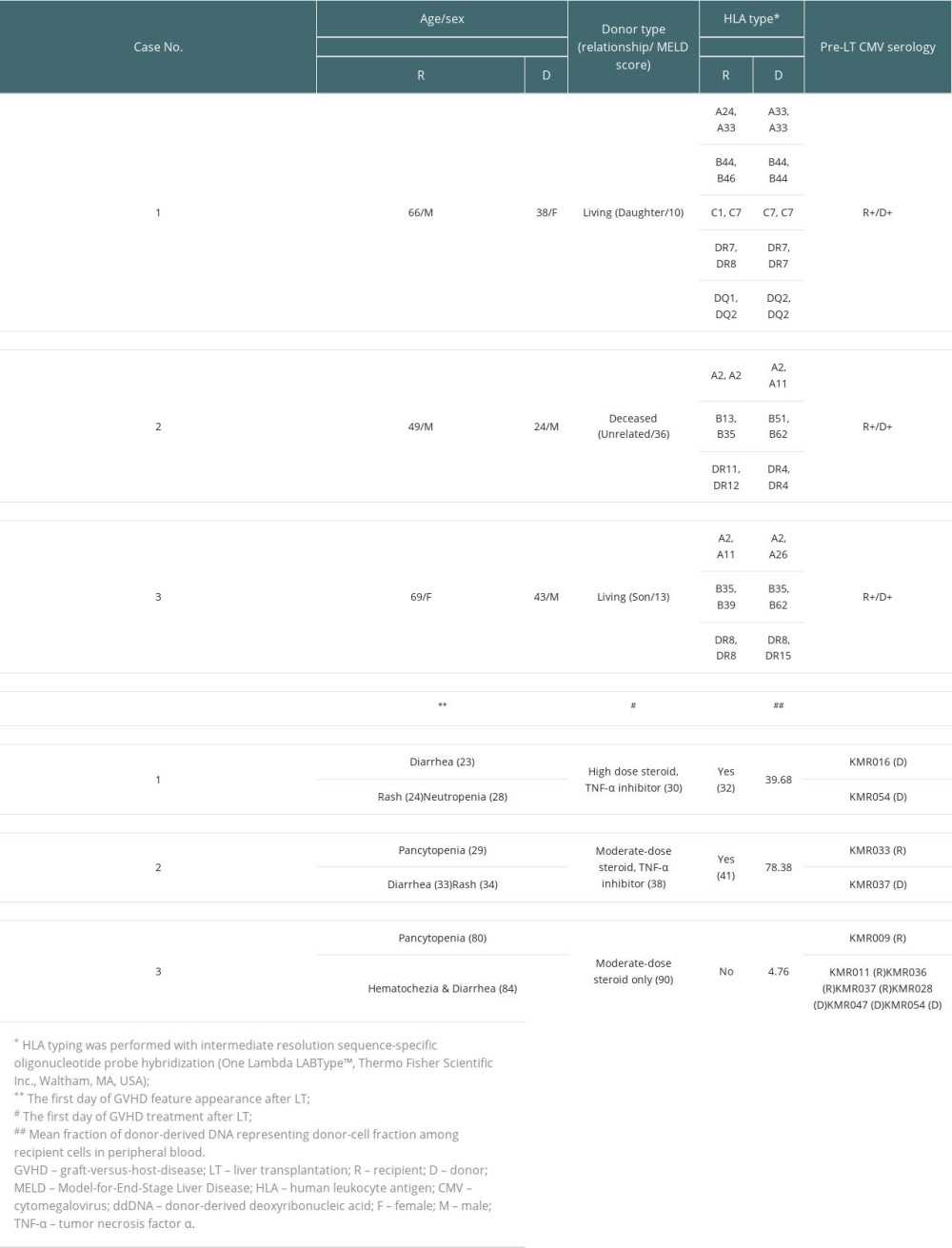 Table 2. Characteristics of patients with graft-versus-host disease after liver transplantation.
Table 2. Characteristics of patients with graft-versus-host disease after liver transplantation.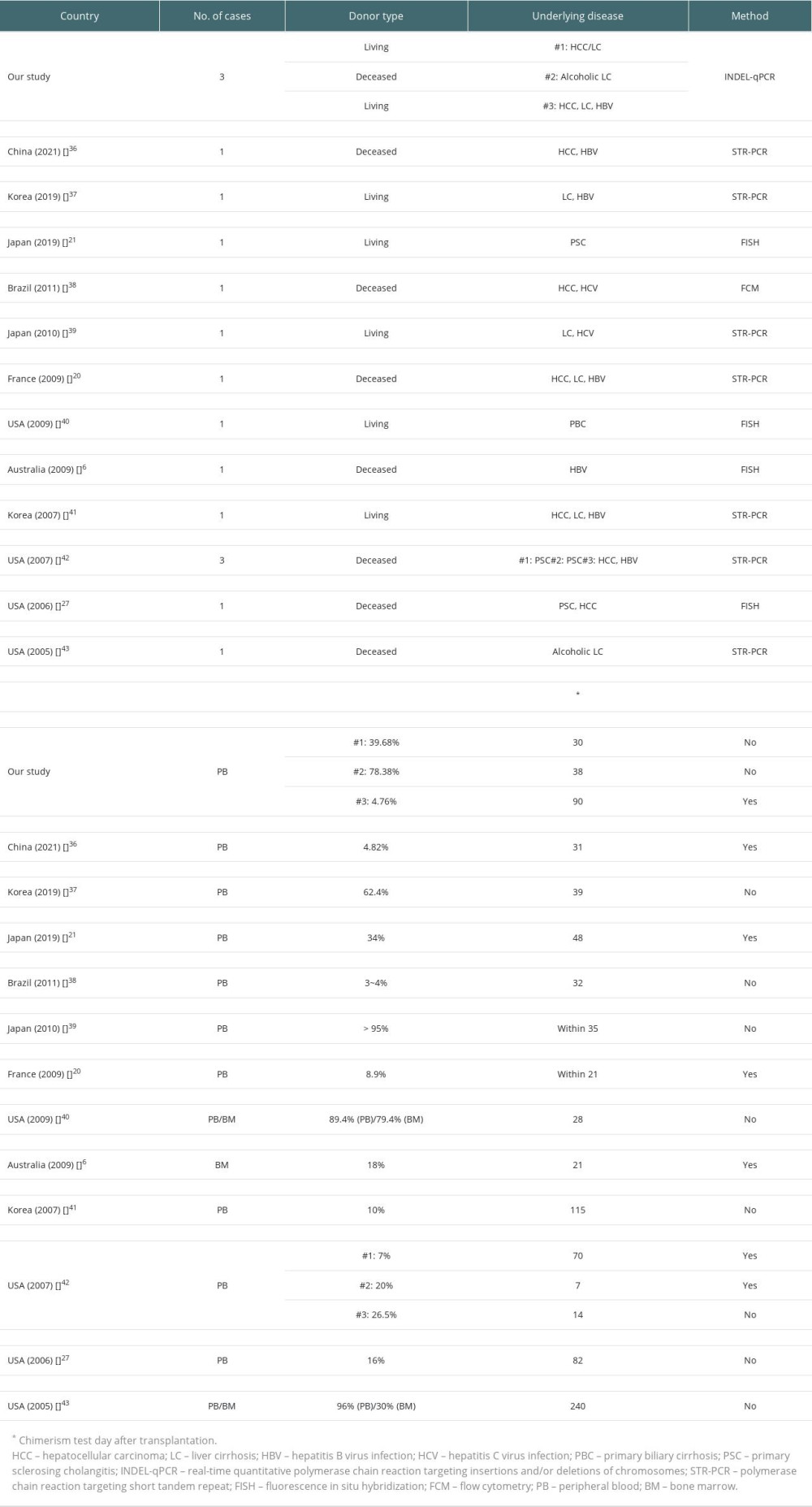 Table 3. Overview of previous chimerism studies of graft-versus-host disease after liver transplantation.
Table 3. Overview of previous chimerism studies of graft-versus-host disease after liver transplantation. Table 1. List of 39 informative markers and their chromosomal locations.
Table 1. List of 39 informative markers and their chromosomal locations. Table 2. Characteristics of patients with graft-versus-host disease after liver transplantation.
Table 2. Characteristics of patients with graft-versus-host disease after liver transplantation. Table 3. Overview of previous chimerism studies of graft-versus-host disease after liver transplantation.
Table 3. Overview of previous chimerism studies of graft-versus-host disease after liver transplantation. In Press
18 Mar 2024 : Original article
Does Antibiotic Use Increase the Risk of Post-Transplantation Diabetes Mellitus? A Retrospective Study of R...Ann Transplant In Press; DOI: 10.12659/AOT.943282
20 Mar 2024 : Original article
Transplant Nephrectomy: A Comparative Study of Timing and Techniques in a Single InstitutionAnn Transplant In Press; DOI: 10.12659/AOT.942252
28 Mar 2024 : Original article
Association Between FEV₁ Decline Rate and Mortality in Long-Term Follow-Up of a 21-Patient Pilot Clinical T...Ann Transplant In Press; DOI: 10.12659/AOT.942823
02 Apr 2024 : Original article
Liver Transplantation from Brain-Dead Donors with Hepatitis B or C in South Korea: A 2014-2020 Korean Organ...Ann Transplant In Press; DOI: 10.12659/AOT.943588
Most Viewed Current Articles
05 Apr 2022 : Original article
Impact of Statins on Hepatocellular Carcinoma Recurrence After Living-Donor Liver TransplantationDOI :10.12659/AOT.935604
Ann Transplant 2022; 27:e935604
12 Jan 2022 : Original article
Risk Factors for Developing BK Virus-Associated Nephropathy: A Single-Center Retrospective Cohort Study of ...DOI :10.12659/AOT.934738
Ann Transplant 2022; 27:e934738
22 Nov 2022 : Original article
Long-Term Effects of Everolimus-Facilitated Tacrolimus Reduction in Living-Donor Liver Transplant Recipient...DOI :10.12659/AOT.937988
Ann Transplant 2022; 27:e937988
15 Mar 2022 : Case report
Combined Liver, Pancreas-Duodenum, and Kidney Transplantation for Patients with Hepatitis B Cirrhosis, Urem...DOI :10.12659/AOT.935860
Ann Transplant 2022; 27:e935860








