22 December 2020: Original Paper
Circulating NKG2A–NKG2D+ CD56dimCD16+ Natural Killer (NK) Cells as Mediators of Functional Immunosurveillance in Kidney Transplant Recipients
Li Zhu12ABCDEF, Hristos Karakizlis3B, Rolf Weimer3B, Christian Morath4B, Naruemol Ekpoom2B, Eman H. Ibrahim2BE, Gerhard Opelz2ADEF, Volker Daniel2ABCDEF*DOI: 10.12659/AOT.925162
Ann Transplant 2020; 25:e925162
Abstract
BACKGROUND: Recently, in patients with long-term functioning allografts, we showed that high NKG2D+ NK cell numbers in the peripheral blood were associated with a higher glomerular filtration rate, whereas high NKG2A+ NK cells were associated with a lower glomerular filtration rate. Both NK cell determinants react with ligands (MIC A/B, HLA-E) expressed on stressed cells, such as virus-infected cells, tumor cells, or cells activated during graft rejection. In the present study, we attempted to characterize these 2 NK cell subsets further.
MATERIAL AND METHODS: Using flow cytometry, NK cell subsets were analyzed in whole-blood samples of 35 stable kidney transplant recipients (serum creatinine mean±SD: 1.44±0.45 mg/dl). Blood was obtained 95–3786 days after transplant (mean±SD: 1168±1011 days after transplant).
RESULTS: High proportions of NKG2A-NKG2D+ NK cells were strongly associated with high numbers of CD56dimCD16+ (p=0.001) NK cells co-expressing CD107 (P=0.001) and granzyme B (P=0.045), suggesting that NKG2A–NKG2D+ NK cells are predominantly cytotoxic. In contrast, high numbers of NKG2A+NKG2D– NK cells were strongly associated with low numbers of CD56dimCD16+ NK cells expressing CD107 (P=0.026), CD25 (p=0.008), TGF-βR (P=0.028), and TGF-β (P=0.005), suggesting that patients with high proportions of NKG2A+NKG2D– NK cells have low proportions of NK cell subsets with cytotoxic phenotype.
CONCLUSIONS: A high proportion of NKG2A+NKG2D– NK cells is associated with decreased counts of NKG2A–NKG2D+ CD56dimCD16+ cytotoxic NK cells in the circulation. This may result in impaired immunosurveillance. We would like to hypothesize that NKG2A–NKG2D+ CD56dimCD16+ cytotoxic NK cells eliminate MIC A/B-expressing stressed cells which possess a potential to harm the transplant. Further studies will have to evaluate whether the proportion of NKG2A–NKG2D+ CD56dimCD16+ cytotoxic NK cells is a useful biomarker for the prediction of an uncomplicated postoperative course in kidney transplant recipients.
Keywords: Blood, Delayed Graft Function, Kidney Transplantation, NK Cell Lectin-Like Receptor Subfamily C, NK Cell Lectin-Like Receptor Subfamily K, Immunologic Surveillance, transplant recipients
Background
NK cells are part of the innate immune system and can eliminate antigens within hours after primary contact, in contrast to the adaptive immune system that needs days for antigen clearance. Thus, NK cells are predestined to mediate immunosurveillance, as proposed by Bléry and Vivier [1] and Lopez-Larrea et al. [2]. Using their NKG2 receptors, they are able to sense stressed cells such as virus-infected or tumor cells [2]. Stressed cells express MIC A/B and/or HLA-E [3]. MIC A/B determinants are ligands for the activating NK cell receptor NKG2D, whereas HLA-E reacts with the inhibiting NK cell receptor NKG2A [3,4]. During acute and chronic rejection of solid allografts, cells express MIC A/B and/or HLA-E [5–7]. We hypothesize that NK cells patrol through the body and eliminate stressed cells at a very early stage of cell activation or cell transformation (e.g., when the number of stressed cells is still very low), and thereby prevent impaired graft function in transplant recipients.
Recently, we showed in renal transplant recipients with long-term functioning allografts that high relative as well as high absolute numbers of circulating NKG2A+NKG2D− NK cells are associated with impaired graft function, whereas high proportions of NKG2D+, particularly NKG2D+ CD56dimCD16+ NK cells, are associated with good function of the transplant [8,9]. The higher the ratio of NKG2A+ CD56dimCD16+ to NKG2D+ CD56dimCD16+ NK cells in the blood, the lower the glomerular filtration rate (GFR) of the transplant [9]. Interestingly, this ratio decreased significantly with time after transplant due to an increase of NKG2D+ CD56dimCD16+ NK cells with prolonged follow-up [9].
We speculated previously that NKG2D+ NK cells might have a protective role in long-term kidney transplant recipients, whereas NKG2A+ NK cells might promote deterioration of graft function [8, 9]. This speculation was prompted by our finding that high numbers of NK cells in the peripheral blood [10], especially of NKG2D+ NK cells [9], were associated with good long-term graft function and with findings of others showing that tolerant kidney and liver transplant recipients displayed an higher proportion of peripheral blood NK cells [11,12]. Our findings are in line with those of Crespo et al., who reported finding a higher serum creatinine level and protein-to-creatinine ratio in urine of patients with donor-specific antibodies (DSA) and higher numbers of NKG2A+ NK cells, compared to those with lower numbers of NKG2A+ NK cell numbers in the blood [13]. Patients with antibody-mediated changes in biopsies showed significantly higher proportions of CD3-CD56+NKG2A+ NK cells than DSA patients without these lesions [13]. The differences appeared unrelated to retransplantation, previous acute rejection, or immunosuppressive therapy [13]. Jung et al. found a higher density of NK cells co-expressing NKG2A in patients with antibody mediated rejection compared to patients with T cell-mediated rejection or no rejection [14]. Kordelas et al. showed that NK cells expressing the activating CD94/NKG2C receptor are present in significantly lower numbers in patients after allogeneic stem cell transplantation with severe acute and chronic graft-versus-host disease (GvHD) [15]. Moreover, the ratio of CD94/NKG2C to CD94/NKG2A was reduced in patients with severe acute and chronic GvHD after receiving an HLA-mismatched graft. The authors concluded that the results provide evidence that CD94/NKG2C is involved in GvHD prevention [15]. Ataya et al. reported that pretransplant adaptive NKG2C+ NK cells protect against CMV infection in kidney transplant recipients [16], and Rohn et al. found that a certain donor MICA allele reactive with NKG2D is a protective prognostic determinant for CMV disease in kidney transplant recipients [17].
Based on these findings, we hypothesize that circulating NK cells with activating NKG2D and/or NKG2C receptors may mediate immunosurveillance, eliminate stressed cells very early after their development, and thereby protect the allograft from rejection. In contrast, NKG2A+ NK cells appear to be harmful for the graft. We therefore thought it of interest to study additional characteristics of NK cells with NKG2A or NKG2D receptors on the cell surface, especially with respect to earlier findings that CD56dimCD16+ NK cells co-expressing CD8, perforin, and granzyme B were associated with high serum creatinine levels and CD56bright (CD56bri) NK cells producing IL10 were associated with low serum creatinine levels, thus subdividing NK cells into cytotoxic and immunoregulatory subsets, with potential clinical relevance [18].
In the present study, we investigated whether NK cells with only NKG2A receptors are associated with NK cell subsets showing opposite characteristics when compared to NK cells with only NKG2D, and whether these are different from those of NK cells with both NKG2A and NKG2D receptors on the cell surface. Furthermore, we examined whether NKG2A+CD56+ NK cells, including both NKG2A+NKG2D− and NKG2A+NKG2D+ cells, are associated with NK cell subsets that are different from those of NKG2D+CD56+ NK cells comprising NKG2A–NKG2D+ as well as NKG2A+NKG2D+ lymphocytes. We investigated whether NKG2A/D is mainly expressed on cytotoxic CD56dimCD16+ and/or on immunoregulatory/cytokineproducing CD56briCD16− NK cell subsets and whether these cells co-express the degranulation marker CD107, perforin, granzyme B, IL17, IFNγ, and/or CD8, the activation maker HLA-DR (HLADR), CD69 and the IL-2 (CD25) and IFNγ (CD119) receptor, the inhibitory cytokines TGFβ, IL10, and IL4, including their surface receptors, and the inhibitory killer cell immunoglobulin-like receptors (KIRs) CD158a, CD158b and CD158e, and whether they are associated with Treg cell numbers in the blood of patients. We wanted to investigate the relationship between NKG2A+ and NKG2D+ NK cells and why they are associated with graft function.
Material and Methods
PATIENTS:
Blood specimens were obtained from 35 kidney graft recipients. Patients were studied during regular visits in the outpatient clinics of the university hospitals Giessen and Heidelberg. Patients were investigated at 95 to 3786 days after transplant (mean±SD: 1168±1011 days after transplant). The study was approved by the Heidelberg Ethics Committee (S-225/2014). Patients gave written informed consent for all assays performed within the study. The study was conducted in adherence to the Declaration of Helsinki. Table 1 summarizes the demographic data of the patients. Transplant recipients received 6 immunosuppressive protocols consisting of the combinations tacrolimus+mycophenolate mofetil (MMF) (n=14), tacrolimus+MMF+steroids (n=10), cyclosporine+MMF+steroids (n=7), steroids+MMF+everolimus (n=2), tacrolimus+MMF+steroids+everolimus (n=1), and tacrolimus+MMF+everolimus (n=1) (Table 1).
LYMPHOCYTE AND NK CELL SUBSETS:
All blood specimens were investigated immediately after arrival in the lab. Surface and intracellular markers were analyzed using immunofluorescence staining. CD45+, CD56+CD16, CD19+, CD3+, CD4+, and CD8+ lymphocyte subsets were routinely determined as previously described [10]. Total CD56+CD16 NK cell count was determined using a mixture of PE-conjugated CD16 and CD56 monoclonal antibodies in the same tube. Multitest CD3/CD16+CD56/CD45/CD19/Trucount (BD Bioscience) is a four-color direct immunofluorescence reagent for enumerating percentages and in combination with Trucount tubes (BD Bioscience) the absolute number of mature human NK cells. In addition, NK cell subsets were studied [18]. We added 150 μl of heparinized whole blood and fluorochrome-labeled monoclonal antibodies against NKG2A, NKG2D, CD158a (KIR2DL1), CD158b (KIR2DL2/L3), CD158e (KIR3DL1), CD8, CD107, CD69, IL2R, IL4R, IL10R, IFNγR, TGFβRII and IgG isotype controls only to certain tubes, whereas antibody against CD45, CD3, CD56, CD16, and HLA-DR was added to each test tube (Table 2). Tubes were vortexed and incubated at room temperature (22°C) in the dark. After 30-min incubation at 22°C, 2 ml lyse solution diluted 1: 10 (BD Bioscience, Sunnyvale, CA, USA) was pipetted in each tube. All tubes were vortexed, incubated for 10 min at room temperature in the dark, and centrifuged for 8 min at 1300 rpm. Supernatants were discarded and 2 ml of PBS was pipetted into each tube. Then, each tube was vortexed and centrifuged for 8 min at 1300 rpm. Supernatants were discarded and pellets were washed again. The cells were suspended in 300 μl of PBS and analyzed with a FACSCanto II triple-laser flow-cytometer (BD Bioscience). When intracellular markers were studied, lymphocytes were permeabilized. We pipetted 500 μl of BD Perm/Wash buffer II diluted 1: 10 (BD Bioscience) to the tubes without Foxp3 monoclonal antibody after the last washing step, and lymphocytes were incubated at room temperature for 10 min. After adding 2 ml of PBS, tubes were vortexed and centrifuged for 8 min at 1300 rpm. Supernatant was removed and discarded. Pellets were suspended in 100 μl of PBS. Antibodies against intracellular determinants IL4, IL10, IL17, TGFβ, IFNγ, perforin, and granzyme B were added. Tubes were incubated at room temperature for 30 min. To the tubes intended for Foxp3 monoclonal antibody, 1000 μl fixation/permeabilization buffer diluted 1: 3 (Foxp3 Stain Buffer Set, eBioscience, Darmstadt, Germany) was pipetted. Then, cells were incubated at room temperature for 30 min, and 2 ml of permeabilization washing buffer diluted 1: 10 (Foxp3 Stain Buffer Set, eBioscience) was added. Lymphocytes were vortexed, centrifuged for 8 min at 1300 rpm, and supernatants were removed and discarded. Antibodies reactive with intracellular determinants such as IFNγ and Foxp3 were added. All tubes were vortexed and incubated at room temperature for 30 min. We then added 2 ml of permeabilization washing buffer (tubes containing Foxp3 antibody) or PBS (tubes without Foxp3 antibody). Then, lymphocytes were washed twice, and, finally, 300 μl of PBS were pipetted to each pellet. Cell numbers were determined with eight-color fluorescence using a FACSCanto II triple-laser flow-cytometer (BD Biosciences). In the initial FSC/SSC dot plot, at least 50 000 lymphocytes were analyzed. Our data reflect the cytokine production of NK cells in vivo because cells were not stimulated for intracellular cytokine staining. Mouse IgG1 isotype controls and monoclonal antibodies were purchased from BD Bioscience, with the exception of anti-NKG2A, −CD158a, −CD158b and −CD158e (Miltenyi Biotec GmbH, Germany) and −TGFβ1 and −TGFβRII (R&D Systems, Inc., Minneapolis, USA). Figure 1 shows the gating strategy for the most relevant NK and Treg cell subsets mentioned in the Results section.
STATISTICAL ANALYSIS:
We studied whether NKG2D and NKG2A were expressed on CD56dimCD16+ and CD56briCD16− NK cells and whether these subsets expressed CD158a, CD158b, CD158e, CD69, CD8, CD107, granzyme B, perforin, TGFβ, IFNγ, IL4, IL10, IL17, IL2R, IFNγR, TGFβR, and HLADR. Cell subsets were compared using the Spearman rank correlation test. Because cell subsets were divided into either lymphocytes with immunoregulatory phenotype (TGFβ+, IL4+, IL10+) or lymphocytes with cytotoxic phenotype (CD8+, CD107+, IFNγ+, perforin+, granzymeB+),
Results
NKG2A+NKG2D− AND NKG2A–NKG2D+ NK CELLS IN THE PERIPHERAL BLOOD:
In the peripheral blood of 35 renal transplant recipients with long-term functioning allografts, 33% of all CD56+ NK cells expressed only NKG2D and 12% only expressed NKG2A, whereas 43% were double-positive for NKG2D and NKG2A (Table 3). Thus, 76% of all circulating NK cells expressed NKG2D and 55% expressed NKG2A.
NKG2A+NKG2D− AND NKG2A–NKG2D+ NK CELLS AND TREG CELLS:
In a previous publication, we showed that high CD56+16 (=CD45+CD3–CD56+CD16+ and CD45+CD3–CD56+CD16−) NK and high CD4+CD25+CD127–Foxp3+ Treg cell numbers were associated with good graft function in patients in the late post-transplant period [10] and that NKG2D+ CD56dimCD16+ NK cells particularly increased in the late post-transplant period [9].
In the present study, absolute CD56+16 NK cell numbers in the late post-transplant period consisted of high proportions of NKG2A–NKG2D+ (r=0.406; P=0.017) and low proportions of NKG2A+ NK cells (r=−0.462; P=0.006), suggesting that most of the peripheral NK cells in transplant recipients with good long-term outcome are NKG2A–NKG2D+, whereas NK cells in patients with impaired graft function are predominantly NKG2A+, as also shown previously [8,9] (Table 4). Moreover, high proportions of CD4+CD25+Foxp3+CD127− Treg were associated with high NKG2D+ (r=0.387; P=0.021) and low NKG2A+NKG2D− NK cells (r=−0.338; P=0.047) (Table 4). Treg and NKG2D+ NK cells might have a synergistic effect on graft function, in contrast to NKG2A+NKG2D− NK cells, which might act antagonistically.
A subset of NK cells, IFNγ+ Treg, which previously have been shown to be associated with good graft function [19,20], were strongly associated with double-positive NKG2A+NKG2D+ NK cells (r=0.740; P=0.004) (Table 4). As recently reported, the particular Treg subset that increases together with total CD56+16 NK cells (CD45+CD3–CD56+CD16+ and CD45+CD3–CD56+CD16− NK cells) expresses the phenotype Helios+IFNγ− and therefore does not produce IFNγ [10]. Our present data are in line with these previous findings and suggest a graft-protective role of NKG2A–NKG2D+ NK cells and Treg cells, independently or in combination with the IFNγ+ Treg subset.
NKG2A+NKG2D− AND NKG2A–NKG2D+ CD56DIMCD16+ NK CELLS:
CD56dimCD16+ is a cytotoxic NK cell phenotype [18]. High proportions of NKG2A–NKG2D+ NK cells were strongly associated with high proportions of CD56dimCD16+ (r=0.541; P=0.001) that were, in addition, CD107+ (r=0.546; p=0.001) and granzyme B+ (r=0.341; P=0.045) (Table 4). The data support the notion that NKG2D+ NK cells indeed are predominantly cytotoxic. In contrast, high proportions of NKG2A+NKG2D− NK cells were strongly associated with low proportions of CD56dimCD16+ that were, in addition, CD107+ (r=−0.377; P=0.026), CD25+ (r=−0.439; P=0.008), TGFβR+ (r=−0.387; P=0.028), and TGFβ+ (r=−0.465; P=0.005). This suggests that patients with high NKG2A+NKG2D− NK cells have low NK cells with an activated cytotoxic CD107+ CD25+ CD56dimCD16+ phenotype and TGFβ production (Table 4). It follows that a domination of NKG2A+NKG2D− NK cells indicates a decrease of NKG2A–NKG2D+ CD56dimCD16+ cytotoxic NK cells in the circulation (r=−0.787; P<0.001), as shown in Figure 2.
NKG2A+NKG2D− AND NKG2A–NKG2D+ CD56BRICD16− NK CELLS:
CD56briCD16− is an immunoregulatory/cytokine-producing NK cell phenotype [18]. High proportions of NKG2A–NKG2D+ NK cells were strongly associated with low proportions of CD56briCD16− NK cells that co-express CD158a (r=−0.388; P=0.021), CD158b (r=−0.413; P=0.014), CD158e (r=−0.444; P=0.008), CD69 (r=−0.561; P<0.001), HLADR (r=−0.460; P=0.005), CD119 (r=−0.693; P<0.001), perforin (r=−0.357; P=0.035), granzyme B (r=−0.354; P=0.037), CD8 (r=−0.417; P=0.014), IL4 (r=−0.369; P=0.029), and/or IL17 (r=−0.401; P=0.019) (Table 4). Obviously, patients with high proportions of NKG2A–NKG2D+ NK cells have low proportions of immunoregulatory and cytokine-producing CD56briCD16− NK cell subsets co-expressing inhibitory CD158a/b/e receptors, CD69 and HLADR activation markers, inhibitory IL4, cytotoxic markers such as perforin, granzyme B, CD8, IL17, and receptors for IFNγ (CD119). NKG2A+NKG2D− NK cells were only associated with high proportions of CD56briCD119+ NK cells (r=0.344; P=0.043) (Table 4).
NKG2A+NKG2D− AND NKG2A–NKG2D+ CD56+ NK CELLS:
When the total CD56+ NK cell pool was analyzed, NKG2A–NKG2D+ NK cells were associated with high proportions of CD107+ (r=0.530; P=0.001), and low proportions of CD119+HLADR+ (r=−0.529; P=0.001) and CD69+HLADR+ (r=−0.492; P=0.003) NK cells, whereas NKG2A+NKG2D− NK cells were associated with high proportions of CD69+HLADR+ (r=0.565; P<0.001) and CD8+HLADR+ (r=0.407; P=0.017) and low proportions of CD107+ (r=−0.433; P=0.009), CD25+ (r=−0.423, P=0.011), CD25+HLADR+ (r=−0.430; P=0.010), IL4–TGFβ+ (r=−0.427; P=0.011), TGFβ+ (r=−0.465; P=0.005), and TGFβR+ (r=−0.349; P=0.050) NK cells (Table 4). The data suggest that NKG2A–NKG2D+ CD56+ NK cells were associated with resting cytotoxic NK cells expressing degranulation marker CD107, whereas NKG2A+NKG2D− CD56+ NK cells were associated with activated NK cell subsets showing low IL2R, CD107, and TGFβ expression.
NKG2A+ AND NKG2D+ NK CELLS:
As shown previously, high proportions of NKG2D+ CD56+ NK cells consisting of NKG2A+NKG2D+ and NKG2A–NKG2D+ NK cells are strongly associated with GFR [8,9]. When the total of NKG2D+ NK cells was analyzed, there were strong associations with high proportions of CD56dimCD16+ NK cells that co-express TGFβ (r=0.520; P=0.001), TGFβR (r=0.462; P=0.008), and CD25 (r=0.459; P=0.006), and with CD56+ NK cells that show the phenotype TGFβ+IL4− (r=0.458; P=0.006), TGFβ+ (r=0.517; P=0.001), TGFβ+HLADR+ (r=0.361; P=0.036) and TGFβR+ (r=0.413; P=0.019), suggesting strong TGFβ production of NKG2D+ CD56dimCD16+ NK cell subsets (Table 4). In contrast, the total of NKG2A+ NK cells was associated with high proportions of CD56briCD16− NK cells that co-express CD69 (r=0.706; P<0.001), HLADR (r=0.490; P=0.003), CD119 (r=0.694; P<0.001), IL17 (r=0.479; P=0.004), IL4 (r=0.400; P=0.017), perforin (r=0.406; p=0.016), granzyme B (r=0.375; P=0.027), no perforin and no granzyme B (r=0.381; P=0.024), CD158a (r=0.355; P=0.037), CD158e (r=0.489; P=0.003) and with high CD56+ NK cells that show the phenotype CD69+HLADR+ (r=0.428; P=0.010) and CD119+HLADR+ (r=0.366; P=0.030). In addition, the total of NKG2A+ NK cells was associated with low proportions of CD56+ NK cells co-expressing CD107 (r=−0.447; P=0.007) and with low CD56dimCD16+ NK cells (r=−0.588; P<0.001) that co-express CD107 (r=−0.513; P=0.002) and granzyme B (r=−0.389; P=0.021) (Table 4). The data suggest a strong activation of NKG2A+ CD56briCD16− NK cells associated with low numbers of resting cytotoxic granzymeB+CD107+ CD56dimCD16+ cells.
Discussion
LIMITATIONS:
Because we were not able to test all markers of NK cell characteristics in a single tube, we studied statistical associations of NKG2A and NKG2D expression with particular NK cell subsets, suggesting a simultaneous increase or decrease of the NK cell subsets or, alternatively, a co-expression of NKG2A/D on a particular subset. Further studies are needed to clarify whether a particular NKG2A+ or NKG2D+ NK cell subset is clinically relevant and can be used as a prognostic biomarker for good long-term graft function.
Conclusions
Our analysis shows that certain NK cell subsets increase or decrease in association with NKG2A/D expression. Based on these data, we conclude that NKG2A–NKG2D+ CD56dimCD16+ NK cells with cytotoxic characteristics circulate through the body and it appears that they eliminate MIC A/B-expressing stressed cells, thereby preventing proliferation of these cells and consequent rejection of the graft. Those patients have low proportions of NKG2A+NKG2D− NK cells in the blood. A domination of NKG2A+NKG2D− NK cells is associated with decreased counts of NKG2A–NKG2D+ CD56dimCD16+ cytotoxic NK cells in the circulation and may result in impaired immunosurveillance. Further studies are needed to evaluate whether high proportions of NKG2A–NKG2D+ CD56dimCD16+ cytotoxic NK cells are a useful biomarker for the prediction of an uncomplicated postoperative course in kidney transplant recipients. NKG2A+NKG2D− NK cells are associated with high proportions of activated cytotoxic CD56briCD16− NK cells, but they co-express the inhibitory receptors CD158a/b/e and NKG2A and might be functionally suppressed and unable to kill cells expressing HLA-E. Furthermore, our data do not provide evidence that NKG2A+ NK cells are associated with IL10+ CD56briCD16− NK cells and harm the graft by downregulation of this immunoregulatory NK cells subset. High proportions of NKG2A+ NK cells might only be an indicator of low proportions of NKG2A–NKG2D+ CD56dimCD16+ cytotoxic NK cells and an impaired immunosurveillance mediated by the latter. NK cells with double positivity for NKG2A and NKG2D have no clinical relevance with respect to long-term graft function. Our data suggest that NKG2A–NKG2D+ CD56dimCD16+ cytotoxic NK cells are the only relevant effector cells that are responsible for the observed associations of NKG2A+ and NKG2D+ NK cells with graft function.
Figures
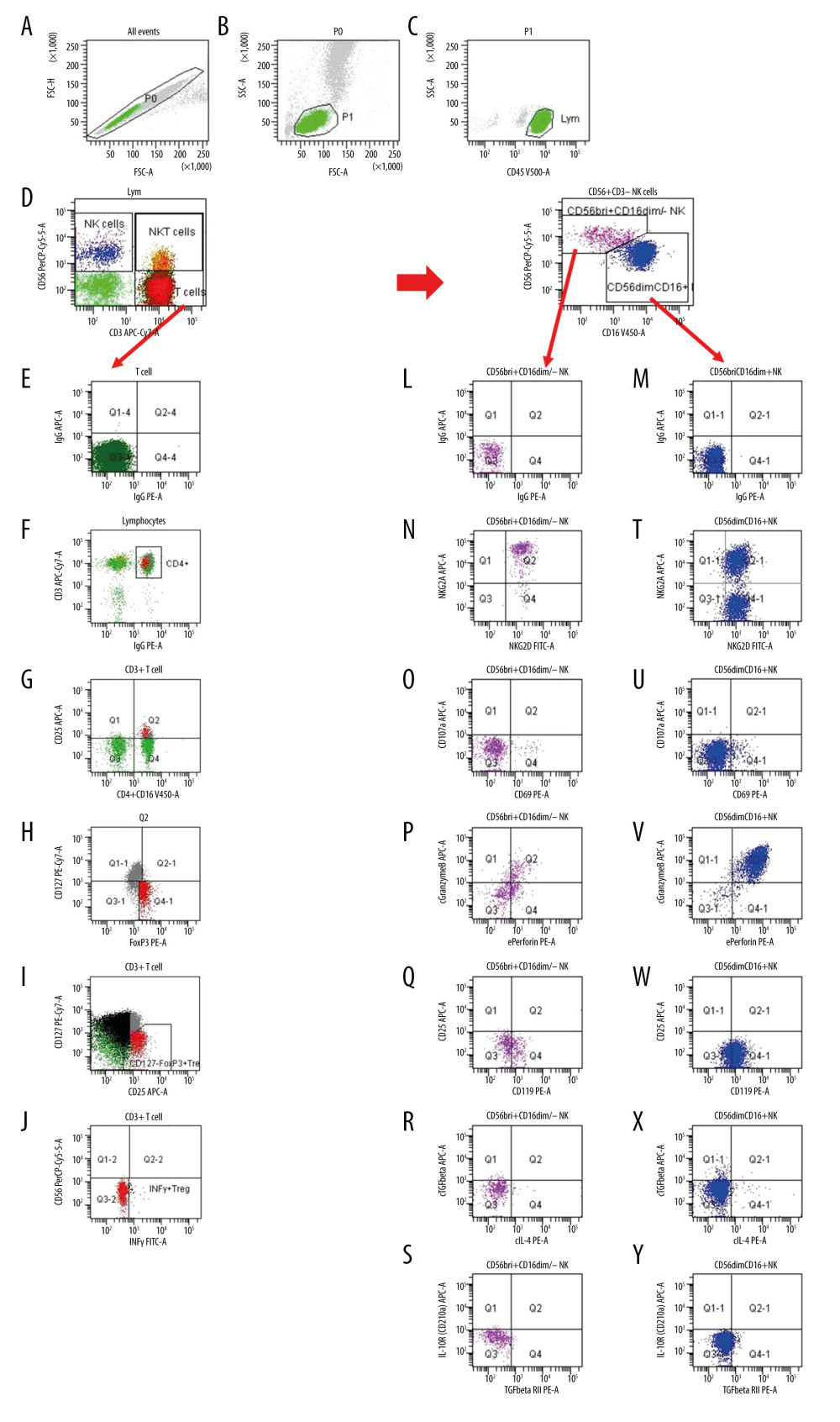 Figure 1. Gating strategy for the determination of NK and Treg cell subsets. (A) After excluding doublets from the total of acquired events, peripheral blood lymphocytes (PBL) were gated according to (B) FSC/SSC and (C) CD45/SSC dot plot. (D) Then, CD3–CD56+ NK cells, CD3+CD56+ NKT cells and CD3+ T cells were gated in the CD3 APC-Cy7/CD56 PerCPCy5.5 dot plot. (K) CD3–CD56+ NK cells were further analyzed according to the intensity of the CD56 and CD16 expression (CD16 V450/CD56 PerCPCy5.5 dot plot). (E, L, M) Further, dependent on isotype controls, subsets of (F–J) T cells, (N–S) CD56brightCD16dim/− NK cells and (T–Y) CD56dimCD16+ NK cells were analyzed using the depicted gate settings in dot plots of (N, T) NKG2D/NKG2A, (O, U) CD69/CD107, (P, V) perforin/granzymeB, (Q, W) IFNγR/CD25, (R, X) IL4/TGFβ and (S, Y) TGFβRII/IL10R. With respect to the determination of IFNγ+ Treg, CD3+ T lymphocytes were further analyzed and (F) CD4+ lymphocytes were identified using a mixture of CD4 and CD16 monoclonal antibody with the same color (CD4+CD16 V450/CD3 APC-Cy7 dot plot). (G) Further, CD4+CD25+ lymphocytes were gated using CD25 and the mixture of CD4 and CD16 monoclonal antibody (CD25 APC/CD4+CD16 V450 dot plot). (H) Then, CD127–Foxp3+ Treg were determined within the CD4+CD25+ lymphocyte subset (CD127 PE-Cy7/Foxp3 PE dot plot). (I) CD127-Foxp3+ Treg were additionally gated in a CD127/CD25 gate based on all CD3+ T cells (CD127 PE-Cy7/CD25 APC dot plot) and (J) IFNγ+ Treg were determined using a CD56/IFNγ dot plot (CD56 PerCPCy5.5/IFNγ FITC). FSC – forward-scattered light; SSC – side-scattered light.
Figure 1. Gating strategy for the determination of NK and Treg cell subsets. (A) After excluding doublets from the total of acquired events, peripheral blood lymphocytes (PBL) were gated according to (B) FSC/SSC and (C) CD45/SSC dot plot. (D) Then, CD3–CD56+ NK cells, CD3+CD56+ NKT cells and CD3+ T cells were gated in the CD3 APC-Cy7/CD56 PerCPCy5.5 dot plot. (K) CD3–CD56+ NK cells were further analyzed according to the intensity of the CD56 and CD16 expression (CD16 V450/CD56 PerCPCy5.5 dot plot). (E, L, M) Further, dependent on isotype controls, subsets of (F–J) T cells, (N–S) CD56brightCD16dim/− NK cells and (T–Y) CD56dimCD16+ NK cells were analyzed using the depicted gate settings in dot plots of (N, T) NKG2D/NKG2A, (O, U) CD69/CD107, (P, V) perforin/granzymeB, (Q, W) IFNγR/CD25, (R, X) IL4/TGFβ and (S, Y) TGFβRII/IL10R. With respect to the determination of IFNγ+ Treg, CD3+ T lymphocytes were further analyzed and (F) CD4+ lymphocytes were identified using a mixture of CD4 and CD16 monoclonal antibody with the same color (CD4+CD16 V450/CD3 APC-Cy7 dot plot). (G) Further, CD4+CD25+ lymphocytes were gated using CD25 and the mixture of CD4 and CD16 monoclonal antibody (CD25 APC/CD4+CD16 V450 dot plot). (H) Then, CD127–Foxp3+ Treg were determined within the CD4+CD25+ lymphocyte subset (CD127 PE-Cy7/Foxp3 PE dot plot). (I) CD127-Foxp3+ Treg were additionally gated in a CD127/CD25 gate based on all CD3+ T cells (CD127 PE-Cy7/CD25 APC dot plot) and (J) IFNγ+ Treg were determined using a CD56/IFNγ dot plot (CD56 PerCPCy5.5/IFNγ FITC). FSC – forward-scattered light; SSC – side-scattered light. 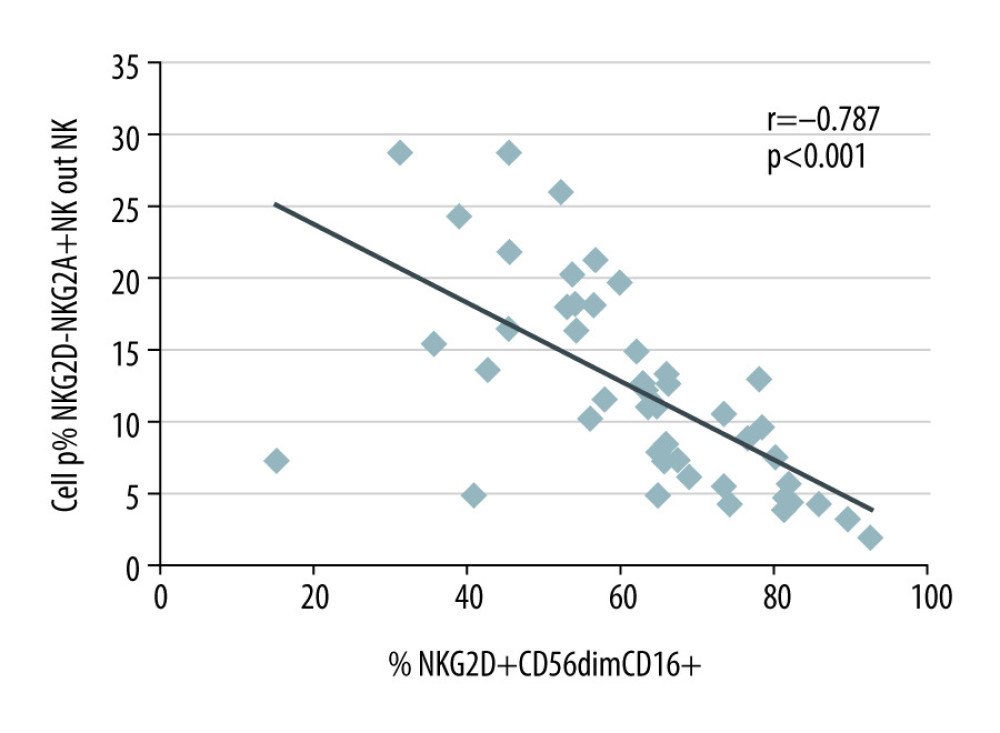 Figure 2. NKG2A+NKG2D− and NKG2D+CD56dimCD16+ NK cells in the blood of 35 kidney transplant recipients with good long-term graft function. NKG2A+NKG2D− NK cells were inversely associated with NKG2D+ CD56dimCD16+ NK cells (r=−0.787; p<0.001).
Figure 2. NKG2A+NKG2D− and NKG2D+CD56dimCD16+ NK cells in the blood of 35 kidney transplant recipients with good long-term graft function. NKG2A+NKG2D− NK cells were inversely associated with NKG2D+ CD56dimCD16+ NK cells (r=−0.787; p<0.001). Tables
Table 1. Demographic data of patients the late post-transplant period. Table 2. Antibody panel for flow cytometric tests of peripheral blood lymphocytes.
Table 2. Antibody panel for flow cytometric tests of peripheral blood lymphocytes.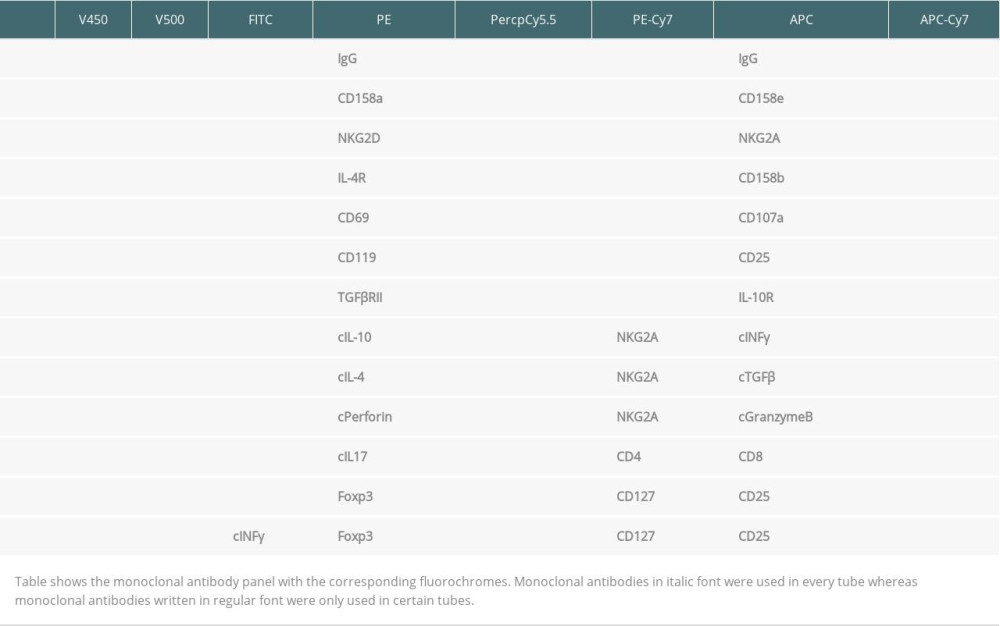 Table 3. Proportion of NKG2A/D-expressing NK cells in the blood of 35 kidney transplant recipients with good long-term graft function.
Table 3. Proportion of NKG2A/D-expressing NK cells in the blood of 35 kidney transplant recipients with good long-term graft function.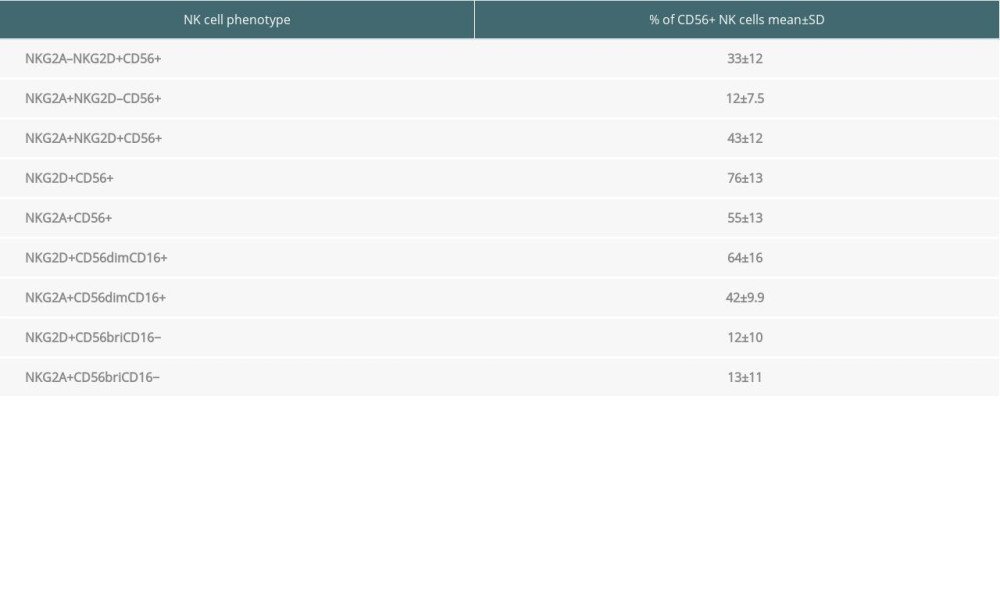 Table 4. Associations of NKG2A/D-expressing NK cells with other NK and T cell subsets in the blood of 35 kidney transplant recipients with good long-term graft function.
Table 4. Associations of NKG2A/D-expressing NK cells with other NK and T cell subsets in the blood of 35 kidney transplant recipients with good long-term graft function.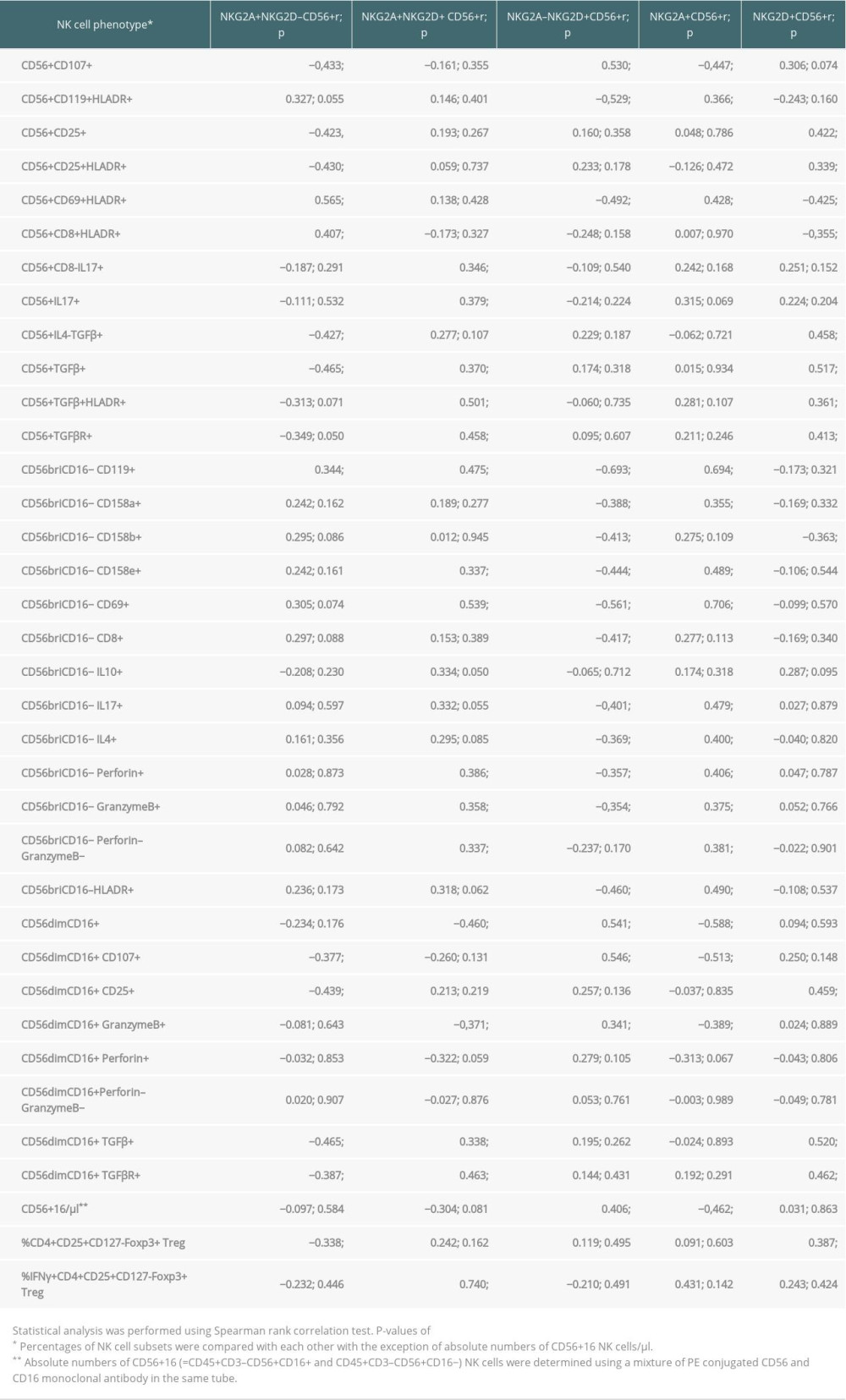
References
1. Blery M, Vivier E, NKG2D-MICA Interaction: A paradigm shift in innate recognition: J Immunol, 2018; 200; 2229-30
2. Lopez-Larrea C, Suarez-Alvarez B, Lopez-Soto A, The NKG2D receptor: Sensing stressed cells: Trends Mol Med, 2008; 14; 179-89
3. Bartel Y, Bauer B, Steinle A, Modulation of NK cell function by genetically coupled C-type lectin-like receptor/ligand pairs encoded in the human natural killer gene complex: Front Immunol, 2013; 4; 362
4. Seliger B, Jasinski-Bergner S, Quandt D, HLA-E expression and its clinical relevance in human renal cell carcinoma: Oncotarget, 2016; 7; 67360-72
5. Zou Y, Stastny P, Susal C, Antibodies against MICA antigens and kidney-transplant rejection: N Engl J Med, 2007; 357; 1293-300
6. Guberina H, Rebmann V, Wagner B, Association of high HLA-E expression during acute cellular rejection and numbers of HLA class I leader peptide mismatches with reduced renal allograft survival: Immunobiology, 2017; 222; 536-43
7. Sullivan LC, Westall GP, Widjaja JM, The presence of HLA-E-restricted, CMV-specific CD8+ T cells in the blood of lung transplant recipients correlates with chronic allograft rejection: PLoS One, 2015; 10; e0135972
8. Zhu L, Aly M, Wang H, Increased natural killer cell subsets with inhibitory cytokines and inhibitory surface receptors in patients with recurrent miscarriage and decreased or normal subsets in kidney transplant recipients late post-transplant: Clin Exp Immunol, 2018; 193; 241-54
9. Zhu L, Aly M, Wang H, Changes of NK cell subsets with time post-transplant in peripheral blood of renal transplant recipients: Transpl Immunol, 2018; 49; 59-71
10. Trojan K, Zhu L, Aly M, Association of peripheral NK cell counts with Helios+ IFN-gamma– Tregs in patients with good long-term renal allograft function: Clin Exp Immunol, 2017; 188; 467-79
11. Sagoo P, Perucha E, Sawitzki B, Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans: J Clin Invest, 2010; 120; 1848-61
12. Newell KA, Turka LA, Tolerance signatures in transplant recipients: Curr Opin Organ Transplant, 2015; 20; 400-5
13. Crespo M, Yelamos J, Redondo D, Circulating NK-cell subsets in renal allograft recipients with anti-HLA donor-specific antibodies: Am J Transplant, 2015; 15; 806-14
14. Jung HR, Kim MJ, Wee YM, CD56(+)CD57(+) infiltrates as the most predominant subset of intragraft natural killer cells in renal transplant biopsies with antibody-mediated rejection: Sci Rep, 2019; 9; 16606
15. Kordelas L, Steckel NK, Horn PA, The activating NKG2C receptor is significantly reduced in NK cells after allogeneic stem cell transplantation in patients with severe graft-versus-host disease: Int J Mol Sci, 2016; 17; 1797
16. Ataya M, Redondo-Pachon D, Llinas-Mallol L, Pretransplant adaptive NKG2C+ NK cells protect against cytomegalovirus infection in kidney transplant recipients: Am J Transplant, 2020; 20; 663-76
17. Rohn H, Tomoya Michita R, Schwich E, The donor major histocompatibility complex class I chain-related molecule A allele rs2596538 G predicts cytomegalovirus viremia in kidney transplant recipients: Front Immunol, 2018; 9; 917
18. Zhu L, Aly M, Wang H, Decreased NK cell immunity in kidney transplant recipients late post-transplant and increased NK-cell immunity in patients with recurrent miscarriage: PLoS One, 2017; 12; e0186349
19. Daniel V, Wang H, Sadeghi M, Opelz G, Interferon-gamma producing regulatory T cells as a diagnostic and therapeutic tool in organ transplantation: Int Rev Immunol, 2014; 33; 195-211
20. Daniel V, Naujokat C, Sadeghi M, Observational support for an immunoregulatory role of CD3+CD4+CD25+IFN-gamma+ blood lymphocytes in kidney transplant recipients with good long-term graft outcome: Transpl Int, 2008; 21; 646-60
21. Lopez-Botet M, Vilches C, Redondo-Pachon D, Dual role of natural killer cells on graft rejection and control of cytomegalovirus infection in renal transplantation: Front Immunol, 2017; 8; 166
22. Daniel V, Trojan K, Adamek M, Opelz G: BMC Immunol, 2015; 16; 45
23. Daniel V, Sadeghi M, Wang H, Opelz G, CD4+CD25+Foxp3+IFN-gamma+ human induced T regulatory cells are induced by interferon-gamma and suppress alloresponses nonspecifically: Hum Immunol, 2011; 72; 699-707
24. Trojan K, Unterrainer C, Weimer R, Helios expression and Foxp3 TSDR methylation of IFNγ+ and IFNγ− Treg from kidney transplant recipients with good long-term graft function: PLoS One, 2017; 12; e0173773
Figures
 Figure 1. Gating strategy for the determination of NK and Treg cell subsets. (A) After excluding doublets from the total of acquired events, peripheral blood lymphocytes (PBL) were gated according to (B) FSC/SSC and (C) CD45/SSC dot plot. (D) Then, CD3–CD56+ NK cells, CD3+CD56+ NKT cells and CD3+ T cells were gated in the CD3 APC-Cy7/CD56 PerCPCy5.5 dot plot. (K) CD3–CD56+ NK cells were further analyzed according to the intensity of the CD56 and CD16 expression (CD16 V450/CD56 PerCPCy5.5 dot plot). (E, L, M) Further, dependent on isotype controls, subsets of (F–J) T cells, (N–S) CD56brightCD16dim/− NK cells and (T–Y) CD56dimCD16+ NK cells were analyzed using the depicted gate settings in dot plots of (N, T) NKG2D/NKG2A, (O, U) CD69/CD107, (P, V) perforin/granzymeB, (Q, W) IFNγR/CD25, (R, X) IL4/TGFβ and (S, Y) TGFβRII/IL10R. With respect to the determination of IFNγ+ Treg, CD3+ T lymphocytes were further analyzed and (F) CD4+ lymphocytes were identified using a mixture of CD4 and CD16 monoclonal antibody with the same color (CD4+CD16 V450/CD3 APC-Cy7 dot plot). (G) Further, CD4+CD25+ lymphocytes were gated using CD25 and the mixture of CD4 and CD16 monoclonal antibody (CD25 APC/CD4+CD16 V450 dot plot). (H) Then, CD127–Foxp3+ Treg were determined within the CD4+CD25+ lymphocyte subset (CD127 PE-Cy7/Foxp3 PE dot plot). (I) CD127-Foxp3+ Treg were additionally gated in a CD127/CD25 gate based on all CD3+ T cells (CD127 PE-Cy7/CD25 APC dot plot) and (J) IFNγ+ Treg were determined using a CD56/IFNγ dot plot (CD56 PerCPCy5.5/IFNγ FITC). FSC – forward-scattered light; SSC – side-scattered light.
Figure 1. Gating strategy for the determination of NK and Treg cell subsets. (A) After excluding doublets from the total of acquired events, peripheral blood lymphocytes (PBL) were gated according to (B) FSC/SSC and (C) CD45/SSC dot plot. (D) Then, CD3–CD56+ NK cells, CD3+CD56+ NKT cells and CD3+ T cells were gated in the CD3 APC-Cy7/CD56 PerCPCy5.5 dot plot. (K) CD3–CD56+ NK cells were further analyzed according to the intensity of the CD56 and CD16 expression (CD16 V450/CD56 PerCPCy5.5 dot plot). (E, L, M) Further, dependent on isotype controls, subsets of (F–J) T cells, (N–S) CD56brightCD16dim/− NK cells and (T–Y) CD56dimCD16+ NK cells were analyzed using the depicted gate settings in dot plots of (N, T) NKG2D/NKG2A, (O, U) CD69/CD107, (P, V) perforin/granzymeB, (Q, W) IFNγR/CD25, (R, X) IL4/TGFβ and (S, Y) TGFβRII/IL10R. With respect to the determination of IFNγ+ Treg, CD3+ T lymphocytes were further analyzed and (F) CD4+ lymphocytes were identified using a mixture of CD4 and CD16 monoclonal antibody with the same color (CD4+CD16 V450/CD3 APC-Cy7 dot plot). (G) Further, CD4+CD25+ lymphocytes were gated using CD25 and the mixture of CD4 and CD16 monoclonal antibody (CD25 APC/CD4+CD16 V450 dot plot). (H) Then, CD127–Foxp3+ Treg were determined within the CD4+CD25+ lymphocyte subset (CD127 PE-Cy7/Foxp3 PE dot plot). (I) CD127-Foxp3+ Treg were additionally gated in a CD127/CD25 gate based on all CD3+ T cells (CD127 PE-Cy7/CD25 APC dot plot) and (J) IFNγ+ Treg were determined using a CD56/IFNγ dot plot (CD56 PerCPCy5.5/IFNγ FITC). FSC – forward-scattered light; SSC – side-scattered light. Figure 2. NKG2A+NKG2D− and NKG2D+CD56dimCD16+ NK cells in the blood of 35 kidney transplant recipients with good long-term graft function. NKG2A+NKG2D− NK cells were inversely associated with NKG2D+ CD56dimCD16+ NK cells (r=−0.787; p<0.001).
Figure 2. NKG2A+NKG2D− and NKG2D+CD56dimCD16+ NK cells in the blood of 35 kidney transplant recipients with good long-term graft function. NKG2A+NKG2D− NK cells were inversely associated with NKG2D+ CD56dimCD16+ NK cells (r=−0.787; p<0.001). Tables
 Table 1. Demographic data of patients the late post-transplant period.
Table 1. Demographic data of patients the late post-transplant period. Table 2. Antibody panel for flow cytometric tests of peripheral blood lymphocytes.
Table 2. Antibody panel for flow cytometric tests of peripheral blood lymphocytes. Table 3. Proportion of NKG2A/D-expressing NK cells in the blood of 35 kidney transplant recipients with good long-term graft function.
Table 3. Proportion of NKG2A/D-expressing NK cells in the blood of 35 kidney transplant recipients with good long-term graft function. Table 4. Associations of NKG2A/D-expressing NK cells with other NK and T cell subsets in the blood of 35 kidney transplant recipients with good long-term graft function.
Table 4. Associations of NKG2A/D-expressing NK cells with other NK and T cell subsets in the blood of 35 kidney transplant recipients with good long-term graft function. Table 1. Demographic data of patients the late post-transplant period.
Table 1. Demographic data of patients the late post-transplant period. Table 2. Antibody panel for flow cytometric tests of peripheral blood lymphocytes.
Table 2. Antibody panel for flow cytometric tests of peripheral blood lymphocytes. Table 3. Proportion of NKG2A/D-expressing NK cells in the blood of 35 kidney transplant recipients with good long-term graft function.
Table 3. Proportion of NKG2A/D-expressing NK cells in the blood of 35 kidney transplant recipients with good long-term graft function. Table 4. Associations of NKG2A/D-expressing NK cells with other NK and T cell subsets in the blood of 35 kidney transplant recipients with good long-term graft function.
Table 4. Associations of NKG2A/D-expressing NK cells with other NK and T cell subsets in the blood of 35 kidney transplant recipients with good long-term graft function. In Press
15 Mar 2024 : Review article
Approaches and Challenges in the Current Management of Cytomegalovirus in Transplant Recipients: Highlighti...Ann Transplant In Press; DOI: 10.12659/AOT.941185
18 Mar 2024 : Original article
Does Antibiotic Use Increase the Risk of Post-Transplantation Diabetes Mellitus? A Retrospective Study of R...Ann Transplant In Press; DOI: 10.12659/AOT.943282
20 Mar 2024 : Original article
Transplant Nephrectomy: A Comparative Study of Timing and Techniques in a Single InstitutionAnn Transplant In Press; DOI: 10.12659/AOT.942252
28 Mar 2024 : Original article
Association Between FEV₁ Decline Rate and Mortality in Long-Term Follow-Up of a 21-Patient Pilot Clinical T...Ann Transplant In Press; DOI: 10.12659/AOT.942823
Most Viewed Current Articles
05 Apr 2022 : Original article
Impact of Statins on Hepatocellular Carcinoma Recurrence After Living-Donor Liver TransplantationDOI :10.12659/AOT.935604
Ann Transplant 2022; 27:e935604
12 Jan 2022 : Original article
Risk Factors for Developing BK Virus-Associated Nephropathy: A Single-Center Retrospective Cohort Study of ...DOI :10.12659/AOT.934738
Ann Transplant 2022; 27:e934738
22 Nov 2022 : Original article
Long-Term Effects of Everolimus-Facilitated Tacrolimus Reduction in Living-Donor Liver Transplant Recipient...DOI :10.12659/AOT.937988
Ann Transplant 2022; 27:e937988
15 Mar 2022 : Case report
Combined Liver, Pancreas-Duodenum, and Kidney Transplantation for Patients with Hepatitis B Cirrhosis, Urem...DOI :10.12659/AOT.935860
Ann Transplant 2022; 27:e935860








