15 August 2023: Review Paper
Free-Circulating Nucleic Acids as Biomarkers in Patients After Solid Organ Transplantation
Joanna Raszeja-WyszomirskaDOI: 10.12659/AOT.939750
Ann Transplant 2023; 28:e939750
Abstract
ABSTRACT: A number types of extracellular DNA (eg, cell-free, cfDNA) circulate in human blood, including mitochondrial, transcriptome, and regulatory DNA, usually at low concentrations. Larger amounts of cfDNA appear in any inflammatory condition, including organ damage due to a variety of reasons. The role of cfDNA in solid organ transplantation is discussed in this review as a valuable additional tool in the standard of care of transplant patients. Post-transplant monitoring requires the use of high-quality biomarkers for early detection of graft damage or rejection to be able to apply early therapeutic intervention. CfDNA complements the traditional monitoring strategies, being a risk stratification tool and an important prognostic marker. However, improving the sensitivity and specificity of cfDNA detection is necessary to facilitate personalized patient management, warranting further research in terms of measurement, test standardization, and storage, processing, and shipping. A diagnostic test (Allosure, CareDx, Inc., Brisbane, CA) for kidney, heart and lung transplant patients is now commercially available, and validation for other organs (eg, liver) is pending. To date, donor-derived cfDNA in combination with other biomarkers appears to be a promising tool in graft rejection as it is minimally invasive, time-sensitive, and cost-effective. However, improvement of sensitivity and specificity is required to facilitate personalized patient management. Whether it could be an alternate to graft biopsy remains unclear.
Keywords: Cell-Free Nucleic Acids, biomarkers, Kidney Transplantation, Liver Transplantation, Heart Transplantation, Pancreas Transplantation, Humans, Organ Transplantation, Tissue Donors, Graft Rejection, DNA
Background
Extracellular DNA was first discovered in human plasma by Mandel and Metais in 1948; there was renewed interest in it in the 1990s and it was recently used in diagnosis of patients [1]. Short circulating nucleic acids are both RNA and DNA [2]. Most come from hematopoietic cells such as leukocytes and are released into the bloodstream during apoptosis and necrosis or through active cell secretion. Human peripheral blood contains, among others, circulating genomic DNA (cfDNA), mitochondrial DNA (cfmtDNA), transcriptome mRNA, and regulatory RNA (eg, miRNA). In healthy people, free nucleic acids come mainly from blood cells and are present in low concentrations. However, with inflammation in the body, larger amounts of free nucleic acids begin to appear in the blood. Characteristic of this phenomenon is the fact that apart from the increased amount, nucleic acids that appear are tissue-specific for the source of the inflammation. Therefore, they can serve as a potential marker of various diseases, indicating a damaged organ [3–5].
Cell-free DNA in the circulating blood plasma of patients with cancer contains tumor-derived DNA sequences that can serve as biomarkers for the early detection of cancers, for guiding therapy, and for monitoring for drug resistance. Abnormal distribution of DNA methylation is a hallmark of many cancers, and methylation changes occur early during carcinogenesis. Systemic analysis of cfDNA methylation profiles, as liquid biopsy, is being developed for early cancer detection, monitoring for minimal residual disease, and predicting treatment response and prognosis, and has emerged as a promising non-invasive diagnostic approach in oncology [6–9].
CfDNA is also currently used in non-invasive prenatal diagnostics as a useful prenatal screening approach for high-quality care of pregnancy. By differentiating the genotype of the mother and the fetus, it is possible to detect changes such as trisomy of chromosome 13, 18, and 21, as well as microdeletions or duplications, starting from 8–10 weeks of pregnancy [10].
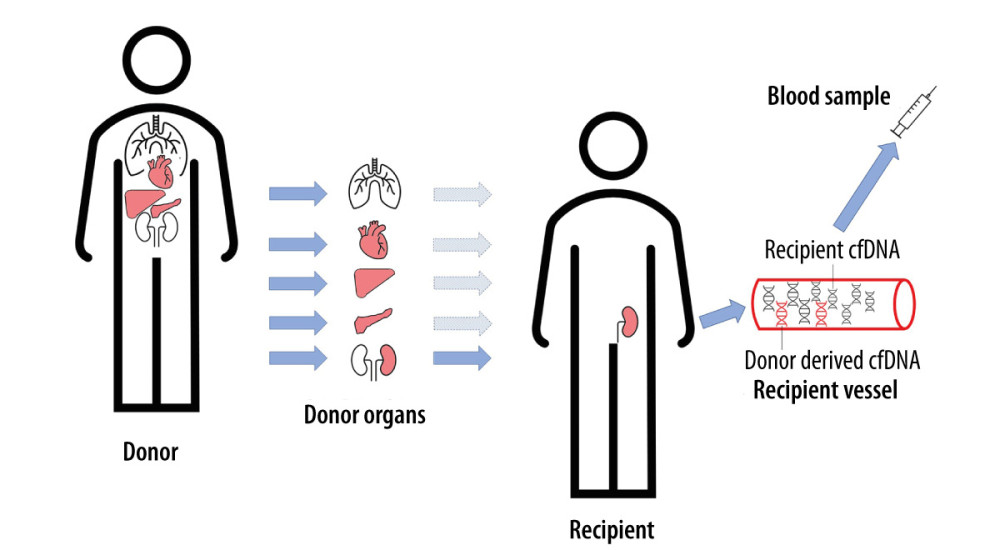
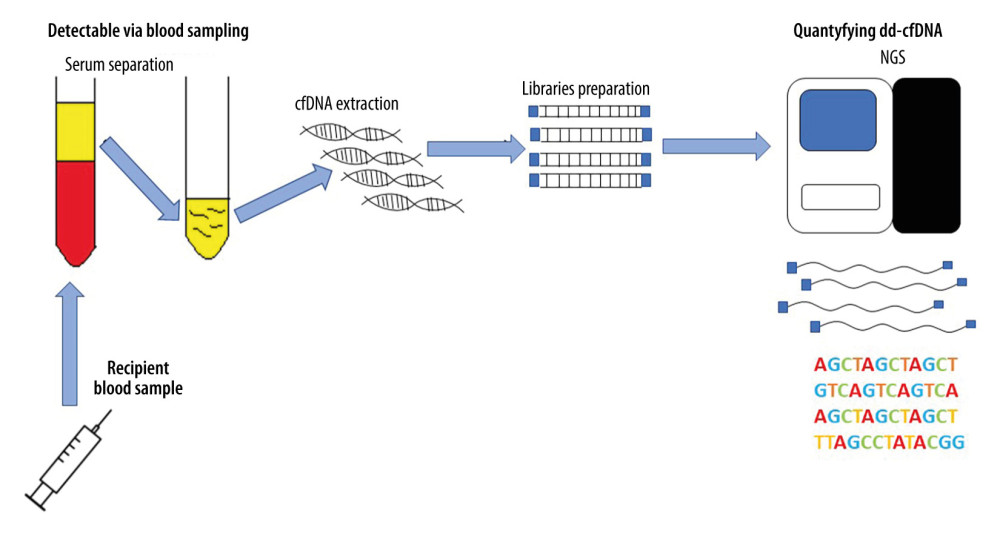
Free-circulating cfDNA can also be used to monitor the health of patients after transplantation [11]. As in the case of pregnant women and cancer patients, a genome different from the patient’s genome will appear in the peripheral blood of transplant recipients. The life-cycle of donor-derived cfDNA (from different organs) is shown in Figure 1. There are many reports in the literature on the monitoring of donor-derived DNA (ddDNA) levels in the entire cfDNA pool of patients after transplantation. The conducted research is based on differences in the genetic information between recipient and donor cfDNAs and on an overall increased amount of cfDNA appearing in the peripheral blood, which is an indicator of ongoing inflammatory changes in the body. If the donor DNA fraction in the pool of slow-circulating DNA increases, it is most likely a sign of transplant rejection. The identification and quantification of cell-free DNA is shown in Figure 2. A diagnostic test for kidney, heart, and lung transplant patients is now commercially available (Allosure, CareDx, Inc., Brisbane, CA). However, the cutoff value is still debatable. CareDx also offers a product called AlloMap for gene expression profiling (GEP).
Liver Transplantation
Liver transplantation (LT) has been saving lives of patients with acute and chronic liver failure since 1963, with very good long-term effects, made possible by progress in surgery, transplant anesthesiology, and hepatology. The long-term success of LT, however, depends on a good balance: adequate suppression of the recipient’s immune system to avoid organ rejection, while maintaining immunosuppression at a level that prevents complications and minimizes adverse effects. The level of immunosuppression required after LT can vary widely among recipients: some patients are highly susceptible to rejection, while others can successfully stop immunosuppression altogether, reaching “operative tolerance”. Despite the judicious use of immunosuppression, up to 27% of LT recipients develop an episode of acute rejection and 68% develop infectious complications [12]. Malignant tumors, chronic renal failure, and complicated metabolic syndrome are more common in liver recipients than in the general population. This shortens the survival time of the patient and the transplanted organ, worsens recipient quality of life, and increases treatment costs. Currently, the standard of care after LT is to add empirical doses of immunosuppressive drugs to recipients, which are then adjusted according to changes in liver function tests (LFT), serum drug levels, or the occurrence of adverse effects. LFTs are an extremely sensitive test for detecting organ damage, but are weakly specific for LT complications, and there are no clear LFT thresholds that are diagnostic or reflect the severity of graft rejection. Similarly, serum calcineurin inhibitor (CNI) concentrations correlate poorly with clinical outcomes in liver recipients, usually involving a series of radiologic and endoscopic examinations culminating in a liver biopsy to diagnose rejection. Not only is this a time-consuming and costly process, but liver biopsies are inherently subjective and invasive. About 1 in 100 leads to serious complications, and 2 in 1000 are fatal.
Cell-free DNA has a short half-life of about 1.5 hours, as these DNA fragments are then rapidly removed from the plasma by the liver, spleen, and kidneys, making it a good marker of “real-time” cell damage because transaminases have a much longer half-life (17 h for AST and 47 h for ALT). While lower levels of circulating free DNA are released during normal physiological turnover, any DNA detected in the circulation reflects cell death (eg, in the graft). Additionally, the levels of detected cell-free DNA are higher in liver and lung recipients compared to kidney or heart recipients, and results from the weight of the transplanted organ [13].
In 7 liver transplant studies, the results clearly showed an increase in detectable levels of graft-derived cell-free DNA prior to clinical manifestation of an episode of biopsy-confirmed acute rejection (BPAR), ranging from 4 to 6 days before elevation of activity transaminases, up to 8–15 days prior to BPAR [11].
A German prospective, observational, multicenter, cohort study showed that extracellular DNA from the liver graft outperformed traditional liver function tests and detects rejection as early as 8–15 days before biopsy-confirmed rejection [14]. The results of the ROC analysis (AUROC 0.97) show a higher diagnostic sensitivity, with 95% specificity for the percentage of cfDNA compared to conventional LFT (AUROC 0.83–0.96). Thus, cfDNA can be used as a „liquid biopsy” for direct and repeated examination of the integrity of the graft and thus for early detection of acute organ rejection, which can help to provide a more timely, effective, and individualized therapy. In patients with infectious complications, the median of donor-specific cell-free DNA was slightly higher than in stable recipients, but lower than in BPAR (5.3–5.7%), and in patients with cholestasis alone this level remained below the 10% cutoff point. Analysis of cfDNA from the graft would therefore be particularly useful when trying to minimize immunosuppressive therapy and in assessing the effectiveness of treatments such as steroid boluses or dose adjustments of immunosuppressive drugs. CfDNA may also be useful for the preselection of an immunosuppressive regimen in patients who require more aggressive therapy because of an increased risk of rejection, or in patients who require less aggressive drug dosing because of the presence of infection, increased risk of infection, or other adverse effects of immunosuppressive therapy [14]. One of the most important conclusions from this study is the finding of a low percentage of chimerism (only 0.068%), which had no effect on the detected cell-free DNA levels. Similar conclusions were reached by researchers from Israel who detected elevated levels of cfDNA from transplanted liver in recipients shortly after transplantation and during an episode of acute graft rejection, as well as in patients after partial hepatectomy and during sepsis, and also described another method of identifying cell-free DNA specifically of hepatic origin [15].
A marginal graft LT recipient has also been described, whose cell-free DNA levels increased rapidly with complications of BPAR, traumatic hematoma, and cytomegalovirus infection, followed by a decrease following successful treatment of each complication [16]. Oellerich et al examined levels of circulating free donor DNA and levels of calcineurin inhibitor (CNI), and found that the lower limit of the therapeutic range of tacrolimus was 8 μg/L, suggesting the usefulness of the test in monitoring liver graft damage in recipients in whom immunosuppression was discontinued [17].
Levitsky et al [18] provided new data regarding the accuracy and negative predictive value of 87% and 100%, respectively, when comparing acute liver graft rejection versus normal graft function. Elevation of donor-derived cfDNA was observed prior to the onset of graft dysfunction and was higher in rejection than dysfunction but no rejection, with decreasing levels after treatment.
Liver transplantation for a selected group of liver malignancies is the only solid organ transplant with high efficacy in curing cancer. There are numerous studies addressing pre- and post-surgery hepatocellular carcinoma (HCC) burden, with a positive association between elevated cfDNA and microvascular invasion. Cancer recurrence and extrahepatic metastases were significantly worse in the cfDNA-positive group of individuals [19–21]. Hann et al [22] published the results of evaluation of urine-derived cfDNA as a potential biomarker of tumor recurrence after treatment. However, extensive studies need to be conducted to evaluate the utility of cfDNA detection in pre- and post-transplant stages in individuals with HCC.
Although graft versus host disease (GvHD) is rare (2%) after liver transplant, has a 75% mortality rate due to non-specific early symptoms, delay in diagnosis, and lack of standard treatment. GvHD occurs due to recognition of host antigens as foreign by immunocompetent T cells in the graft, followed by extensive tissue damage and life-threatening complications. A pilot study by Ha et al [23] showed the promising role of early detection of donor cell chimerism using peripheral blood donor-derived DNA through real-time PCR reaction targeting 39 insertion and/or deletions of chromosomes to halt fatal progression of GvHD after liver grafting.
Pancreas Transplantation
Although few pancreas transplantations are performed, they are increasing. In this kind of transplantation, the risk of acute rejection is estimated at 20%, with a high risk of graft loss after rejection. The risk of pancreas transplantation is also higher compared to other organs, indicating the need for better biomarkers for early detection of graft rejection. There are a few published studies regarding using cell-free DNA in pancreas/kidney transplant recipients (SPKT). Gadi et al found that detection of cfDNA was high in the first month after pancreas transplantation, with a rapid decline thereafter, and with increased correlation with biopsy-proven acute allograft rejection [24]. In another study, Williams et al [25] found that levels of circulating donor cell-free DNA are significantly lower in stable recipients and even those with pancreatitis compared to patients who experienced acute rejection. Researchers in Barcelona explored donor circulating DNA as a diagnostic biomarker before biopsy, finding that assessment of donor-derived DNA 45 days after transplant had the highest specificity and sensitivity to predict acute rejection in biopsy results compared to typical markers like amylase and lipase [26]. Similarly, multicenter study including SPKT and pancreas transplantation alone (PTA) showed that dd-cfDNA is elevated within 3 months after transplant, regardless of the type of transplantation and presence of infection or rejection [27]. On the other hand, a poster presentation at the 2019 American Transplant Congress showed significantly higher median cfDNA in SPKT with graft dysfunction, presence of DSA, or biopsy-proven graft rejection, when compared to stable patients [28]. Additionally, a poster by Ali et al [29] presented at the 29th International Congress of the Transplantation Society (TTS 2022) showed utilization of dd-cfDNA in simultaneous pancreas and kidney transplantation (SPK). After discharge, 16 recipients had donated blood for the analysis of donor-derived DNA together with amylase, lipase, DSA, and BKV (Poliomavirus) PCR. Asymptomatic variability was observed in first 2 months, explained as a resolution of ischemic-reperfusion injury. For up to 2 years, 4 patients had 6 immunological events defined as suspected allograft rejection or subtherapeutic immunosuppression. Compared to 12 stable recipients, those 4 patients had significantly higher donor-derived cfDNA levels.
Heart Transplantation
Both forms of acute transplanted heart (HTx) rejection – acute cellular rejection (ACR) and antibody-mediated rejection (AMR) – can result in graft dysfunction and failure with hemodynamic compromise and mortality. Non-invasive methods like echocardiography or MRI are considered to be insufficiently sensitive and specific to reliably detect rejection; therefore, endomyocardial biopsy is the current criterion standard, but it has well-known limitations. cfDNA was shown to decline 7 days after HTx, and remained consistently low [30], but with gradual increase in median dd-cfDNA levels 2 years after HTx. Moreover, several assays that quantify the level of donor-derived cfDNA after HTx showed an increase in the dd-cfDNA fraction in plasma, which was correlated with acute graft rejection [9,31]. The results of a 26-center study with 740 patients showed 44% sensitivity to detect rejection and a 97% negative predictive value of cfDNA at a 0.2% threshold [32]. Kim et al [33] examined the performance characteristics of a novel test for detecting AR in adult HTx recipients. They obtained plasma samples with contemporaneous EMBs obtained from 223 HTx recipients. They found that the percentage of dd-cfDNA was higher in AR (median 0.58%, IQR, 0.13–1.68%) compared to non-AR (median 0.04%, IQR, 0.01–0.11%, pc<0.001). Increased dd-cfDNA can precede histopathological changes on endomyocardial biopsy as well as subsequent allograft vasculopathy and reduced left ventricular ejection fraction. They concluded that this novel dd-cfDNA test detects AR in HTx recipients with good accuracy – the positive and negative predictive values were 25.1% (95% CI, 18.8–31.5%) and 97.3% (95% CI, 95.1–99.5%), respectively – and holds promise as a non-invasive test for AR in HTx recipients. Thus, cfDNA has emerged as a promising non-invasive tool that can change the approach to rejection surveillance.
As the COVID-19 pandemic has reduced access to endomyocardial biopsy (EMB) rejection surveillance in HTx, Amadio et al [34] performed non-invasive rejection surveillance with gene expression profiling (GEP) and donor-derived cell-free DNA (dd-cfDNA) testing in 90 HTx recipients more than 6 months after transplantation. All 4 patients with a positive dd-cfDNA result (range: 0.19–0.81%) underwent EMB with no significant cellular or antibody-mediated rejection. Consequently, 15 cases (42%) had reduction in immunosuppression, and this increased to 55% in HTx recipients with negative concordant testing. In addition, this non-invasive rejection surveillance could reduce immunosuppression and the HT patients were more satisfied and less anxious. It was also of utmost importance during the pandemic to reduce the exposure for patients and healthcare providers.
Rodgers et al [35] compared the testing accuracy for AR of 2 commercially available dd-cfDNA and gene expression profiling (GEP) tests in 112 HTx recipients. They found a very good positive correlation between standard SNP and expanded SNP assays performed in 428 samples (
Similarly, Henricksen et al [36] tested eligibility for rejection surveillance with GEP (n=95) in comparison with a paired testing cohort (n=64) and surveillance from both dd-cfDNA and GEP. They found that pairing dd-cfDNA and GEP testing yielded similar survival and 1-year rejection-free survival, while requiring significantly fewer endomyocardial biopsies. To date, prospective controlled studies addressing clinical utility of absolute dd-cfDNA concentration versus% dd-cfDNA are lacking.
Lung Transplantation
Lung transplantation is burdened by limited long-term survival, mainly by the development of bronchiolitis obliterans syndrome (BOS), which is a subset of chronic lung allograft dysfunction (CLAD). Irreversible lung damage is already present at the time of diagnosis; therefore, a useful biomarker for allograft damage is needed.
The CfDNA fraction showed a similar pattern of rapid decline within the first week after transplantation and elevation in patients with acute graft rejection. The results of a multicenter study with 104 participants revealed high variability of cfDNA levels, and patients with the highest levels had a 6.6-fold higher risk of allograft failure, although more than half of them did not have clinical complications. This may be the result of differences in tissue mass (single versus bilateral lung transplant) and rates of cellular turnover [37]. Improved technical development has made it possible to differentiate donor-derived cfDNA from recipient-derived cdDNA with further quantification of each portion using targeted preamplification and droplet digital polymerase chain reaction. The donor fraction has been associated with graft injury of transplanted kidneys [3], livers [14], hearts [38] and lungs [37]. A pilot study by Magnusson et al showed that analysis of the quantity and relative proportion of dd-cfDNA and recipient-derived cfDNA might be a promising method of detecting CLAD [39].
Kidney Transplantation
In kidney transplant recipients, routine monitoring with cfDNA has been reported to accurately identify and characterize graft injury, correlate with pathologic findings, and assess response to therapy, including treatment of rejection [40–44]. Even more importantly, elevation of cfDNA preceded clinically apparent organ injury in solid organ transplantation [45]. While the effectiveness of cfDNA use has been established in clinical trials, its utility in routine clinical practice has not been well described. In 2022, the results of The ADMIRAL study (Assessing AlloSure Dd-cf-DNA, Monitoring Insights of Renal Allografts with Longitudinal Surveillance; NCT04566055) – a large, multicenter, observational, cohort study of 1092 kidney transplant recipients monitored with dd-cf-DNA for ≤3 years – were published [46]. The study validated clinical trial data by documenting the effectiveness of cfDNA use in identifying allograft rejection and subclinical changes in a real-world setting and evaluated the relationship between cfDNA measurements and nonimmune allograft injury. It also assessed the relationship between elevation in cfDNA and important predictors of long-term graft survival, including estimated glomerular filtration rate (eGFR) and formation of
Gray et al [47] performed an interventional study to learn how cfDNA can be used in clinical practice regarding immunosuppression, management of infection, treatment of various types of rejection, and to control or even prevent the formation of dnDSAs. Dandamudi et al [48], in the November 2022 issue of Clinical Journal of American Society of Nephrology, published data on 290 plasma samples stored at a prospective biobank at a St Louis center, collected monthly from 57 children in the first year after kidney transplant between January 2013 and December 2019. They found that cfDNA levels in children remained persistently elevated for at least 4 months after transplantation. In case of a greater disparity in size between the donor and the recipient, they were elevated for up to 1 year after transplantation, before reaching a steady low level. They used a 1% cutoff point for donor-derived cfDNA to discriminate biopsy-proven acute rejection from no rejection, with a receiver operating characteristic area under the curve of 0.82. This cutoff led to a high 96% specificity but low 33% sensitivity. They also concluded that the presence of BK viruria or viremia resulted in a higher median donor-derived cfDNA than before or after the infection. In addition, they used a cutoff >0.5% to predict a wider spread in the eGFR over the next 30 days but not the 1-year outcomes. In December’s 2022 issue of Transplantation, Halloran et al [49] analyzed 367 sequential indication biopsy samples out of 426 collected from the Trifecta study (ClinicalTrials.gov # NCT04239703). They found that combination of donor-derived cfDNA fraction and quantity was significantly more predictive than either variable alone. They used a ≥1% dd-cfDNA cutoff or the ≥78 cp/mL quantity dd-cfDNA cutoff as rejection, or for clinical purposes, as “at-risk for rejection”. They also validated a 2-threshold algorithm for discriminating AR from non-AR using cfDNA in KTx patients. This new 2-threshold algorithm had a sensitivity of 83.1%, specificity of 81.0%, PPV of 67.8%, and NPV of 90.8%, with a 32.6% prevalence of AR in the test set.
Many abstracts presented during the 29th International Congress of the Transplantation Society (TTS 2022) addressed this issue. Tedesco et al [50] assessed outcomes of the QSant test as a diagnostic tool to differentiate between alloimmune injury (AI) and acute rejection (AR). QSant (https://nephrosant.com/products/qsant/) is a urine-based test, which quantifies AR by analyzing the following biomarkers: cfDNA, methylated cfDNA, proteinuria, CXCL10, clusterin (apoliprotein J), and creatinine. The study group included 65 kidney recipients who donated 431 urine samples during 3 months after transplantation. In 57% of patients, DGF was observed. Protocol biopsies were performed in 92% of patients, of which 12% had AR. When compared to results of the QSant test, cfDNA was the most important significant factor determining whether clinical symptoms are the result of AI or AR. Among 21 patients with no-AR in biopsy, 1/3 had QSant >55 and clinical symptoms of AR, suggesting that the biopsies were missed. Results of QSant had proven recovery from IRI in 90% of patients during 3-month follow-up. The authors proposed using QSant for monitoring DGF and for determination of the need to perform a biopsy.
Allam et al [51] compared the results of histologic biopsies with a non-rejection diagnosis or tubular injury and level of serum donor-derived cfDNA (dd-cfDNA) from the AlloSure Registry. The data were collected from recipients in the first month after transplant and compared with results at the end of the first year. There was no difference among the levels of dd-cfDNA, biopsies with no rejection, and outcomes at 1 year. These results suggested that dd-cfDNA identified with non-action histological findings can help make decisions and avoid unnecessary biopsies. Weir et al [52] performed a multi-center study, in which they compared results from 65 biopsies taken from 59 patients who had elevated dd-cfDNA levels, and 540 biopsies obtained due to other causes. A significantly higher incidence of AR was found in patients with elevated dd-cfDNA and significantly lower incidence of AI compared to the other group, suggesting that measuring dd-cfDNA can help make decisions on the need to perform a biopsy. However, Shibab et al [53] assessed the dd-cfDNA concentration in recipients with histological diagnosis of a borderline T cell-mediated rejection (BL-TCLMR). In the Kidney Allograft Outcomes AlloSure Registry, 56 cases of BL-TCMLR were identified. Due to heterogeneity in the group among individual combinations of tubulitis and inflammation, no differences or trajectory was observed regarding concentrations of dd-cfDNA. Obrisca et al [54] observed 49 kidney recipients during at least 1-year follow-up, showing that despite initial stable graft function with creatinine eGFR 1.64±0.58 mg/dl and eGFR 48±21 ml/min/1.73 m2, patients with elevated dd-cfDNA had faster deterioration of kidney function at a mean 15-month follow-up. In a multivariate analysis, dd-cfDNA was an independent risk factor for percent decline of eGFR. Guo et al [55] assessed the prognostic value of dd-cfDNA in a 5-year follow-up and created a dynamic model of graft survival.
Ramirez-Bajo et al [56] combined dd-cfDNA and gene expression assay to improve diagnosis of subclinical acute rejection in simultaneous pancreas and kidney transplantation in a study performed in 77 recipients. Plasma samples were collected before Tx and postoperatively (1 h, 24 h, 7 days, 3 weeks, and after 12 months after transplant), together with protocol biopsies. Dd-cfDNA was assessed using Genoma AlloNex, and gene expression profiles were analyzed with TruGraf using a PCR-based assay. Results were assigned as rejection or non-rejection. In the first measurements, peaks of dd-cfDNA were observed due to ischemia-reperfusion injury. Patients with proven AR–TCMR (T cell-mediated rejection) or ABMR (antibody-mediated rejection) (the latter appeared significantly often), had significantly higher concentrations of dd-cfDNA compared to those with no proven AR in biopsy. A 1% threshold of dd-cfDNA had 86% predictive value to detect rejection, whereas the negative predictive value was 69%.
Miles et al [57] presented a case of pregnant woman after living-donor KTx with stable graft function. During pregnancy, levels of dd-cfDNA were stable and comparable to pre-pregnancy, until a rapid increase during an episode of pre-eclampsia, prompting delivery at 31 weeks. The post-delivery period showed a rapid decay of the graft. The authors suggested that monitoring dd-cfDNA could serve as a novel approach for allograft surveillance in such high-risk patients.
A research group from Ljubljana assessed the utility of analyzing the appearance and cargo (EV-DNA) of urinary extracellular vesicles (uEVs) and cfDNA with respect to incidence of AI and AR in KTx [58]. They collected 40-second morning urine samples before protocol or for-cause kidney biopsy to assess uEV-DNA and cfDNA. The sizes of uEVs in the AI and AR groups compared to stable patients differed significantly (177.5 nm and 174.1nm vs 160.7 nm, respectively). Levels of creatinine EV-DNA copies/mmol differed significantly between stable kidneys and AR, as well as between stable kidneys and AR. CfDNA copies/mmol of creatine in urine differed significantly between stable and AR kidneys. The analysis showed that uEVs, EV-DNA, and cfDNA in urine are a good diagnostic tool for assessment of allograft function and type of injury.
Summary
Cell-free DNA was studied in a variety of recipients, and the vast majority of the data came from plasma assays, with different technology platforms used. Data are presented in Table 1. In addition, on the website clinicaltrials.gov, 31 studies were found, but only 9 are recruiting patients. They are presented in detail in Table 2.
Limitations
The effectiveness of cfDNA use has been established in clinical trials, but utilizing cfDNA as a transplant biomarker needs further investigation in terms of measuring the absolute amount of dd-cfDNA, or% of dd-cfDNA or their combination, and standardizing the assays, as well storage, processing, and shipping. It also has to be taken into account that plasma dd-cfDNA, as reported as a percentage, represents the fraction of donor-to-donor plus recipient cell-free DNA and thus is also influenced by the amount of recipient cell-free DNA in the plasma, and the cutoff point still needs to be determined. Another limitation is that elevated dd-cfDNA did not discriminate between alloimmune and non-alloimmune causes of injury; therefore, there is a need for validation. In general, dd-cfDNA has poor specificity to distinguish between specific causes of allograft injury. Thus, without corresponding histopathology, dd-cfDNA alone cannot reliably differentiate between acute cellular rejection and antibody-mediated rejection. In addition, in transplant recipients, drug toxicity, sepsis, and other complications are common and can trigger injury to the recipients’ organs, causing a rise in recipient-derived cfDNA, and an erroneously low plasma percentage of dd-cfDNA, even in the presence of concurrent dysfunction/injury of the allograft. There are no data on the use of dd-cfDNA in multiorgan transplant recipients. DD-cfDNA quantification may prove to be valuable, but combining dd-cfDNA with other biomarkers will improve the diagnostic potential [78]. Post-transplant monitoring requires high-quality biomarkers for early detection of graft damage or rejection and early therapeutic intervention. Increased dd-cfDNA has been associated with graft rejection, and its assessment is minimally invasive, quick, and cost-effective. In a pediatric study [48], despite high specificity, the sensitivity was low, while in adults this was overcome by developing an algorithm [49]. Therefore, improvement of sensitivity and specificity is required to facilitate personalized patient management [78].
Conclusions
Existing evidence shows that dd-cfDNA is a safe, convenient, and reliable method of acute rejection monitoring in solid organ transplant recipients. Donor-derived cfDNA appears to realize our quest for the development of precision medicine techniques, having potential as a surveillance monitoring tool, and screening patients who would most benefit from proceeding to biopsy. In addition, taking into account shortages of pathologists experienced in assessment of graft biopsies, as well as the crucial time to obtain the assessment (but hopefully without a new pandemic), liquid biopsy in the form of a dd-cfDNA assay is a promising future alternative.
References
1. Zhou M, Hara H, Dai Y, Circulating organ-specific microRNAs serve as biomarkers in organ-specific diseases: Implications for organ allo- and xeno-transplantation: Int J Mol Sci, 2016; 17(8); 1232
2. Wei L, Gong X, Martinez OM, Krams SM, Differential expression and functions of microRNAs in liver transplantation and potential use as non-invasive biomarkers: Transpl Immunol, 2013; 29(1–4); 123-29
3. Gielis EM, Beirnaert C, Dendooven A, Plasma donor-derived cell-free DNA kinetics after kidney transplantation using a single tube multiplex PCR assay: PLoS One, 2018; 13(12); e0208207
4. Zhao D, Zhou T, Luo Y, Preliminary clinical experience applying donor-derived cell-free DNA to discern rejection in pediatric liver transplant recipients: Sci Rep, 2021; 11(1); 1138
5. Zhu X, Ng HI, Xuan L, Sequencing data of cell-free DNA fragments in living-related liver transplantation for inborn errors of metabolism: Data Brief, 2020; 29; 105183
6. Jamshidi A, Liu MC, Klein EA, Evaluation of cell-free DNA approaches for multi-cancer early detection: Cancer Cell, 2022; 40(12); 1537-49e12
7. Lo YMD, Han DSC, Jiang P, Chiu RWK, Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies: Science, 2021; 372(6538); eaaw3616
8. Luo H, Wei W, Ye Z, Liquid biopsy of methylation biomarkers in cell-free DNA: Trends Mol Med, 2021; 27(5); 482-500
9. Snyder TM, Khush KK, Valantine HA, Quake SR, Universal noninvasive detection of solid organ transplant rejection: Proc Natl Acad Sci USA, 2011; 108(15); 6229-34
10. Wang JW, Lyu YN, Qiao B, Cell-free fetal DNA testing and its correlation with prenatal indications: BMC Pregnancy Childbirth, 2021; 21(1); 585
11. Knight SR, Thorne A, Lo Faro ML, Donor-specific cell-free DNA as a biomarker in solid organ transplantation. A systematic review: Transplantation, 2019; 103(2); 273-83
12. Charlton M, Levitsky J, Aqel B, International Liver Transplantation Society consensus statement on immunosuppression in liver transplant recipients: Transplantation, 2018; 102(5); 727-43
13. Beck J, Bierau S, Balzer S, Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury: Clin Chem, 2013; 59(12); 1732-41
14. Schutz E, Fischer A, Beck J, Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study: PLoS Med, 2017; 14(4); e1002286
15. Lehmann-Werman R, Magenheim J, Moss J, Monitoring liver damage using hepatocyte-specific methylation markers in cell-free circulating DNA: JCI Insight, 2018; 3(12); e120687
16. Kanzow P, Kollmar O, Schutz E, Graft-derived cell-free DNA as an early organ integrity biomarker after transplantation of a marginal HELLP syndrome donor liver: Transplantation, 2014; 98(5); e43-45
17. Oellerich M, Shipkova M, Asendorf T, Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study: Am J Transplant, 2019; 19(11); 3087-99
18. Levitsky J, Kandpal M, Guo K, Donor-derived cell-free DNA levels predict graft injury in liver transplant recipients: Am J Transplant, 2022; 22(2); 532-40
19. Long G, Fang T, Su W, The prognostic value of postoperative circulating cell-free DNA in operable hepatocellular carcinoma: Scand J Gastroenterol, 2020; 55(12); 1441-46
20. Ono A, Fujimoto A, Yamamoto Y, Circulating tumor DNA analysis for liver cancers and its usefulness as a liquid biopsy: Cell Mol Gastroenterol Hepatol, 2015; 1(5); 516-34
21. Wang J, Huang A, Wang YP, Circulating tumor DNA correlates with microvascular invasion and predicts tumor recurrence of hepatocellular carcinoma: Ann Transl Med, 2020; 8(5); 237
22. Hann HW, Jain S, Park G, Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma: Hepatoma Res, 2017; 3; 105-11
23. Ha C, Kim SJ, Kim JM, Detecting donor-derived DNA by real-rime PCR in recipients suspected of graft-versus-host-diseases after liver transplantation: A case series and literature review: Ann Transplant, 2023; 28; e938287
24. Williams MD, Fei M, Schadde E, Early experience using donor-derived cell-free DNA for surveillance of rejection following simultaneous pancreas and kidney transplantation: Transplant Direct, 2022; 8(5); e1321
25. Ventura-Aguiar P, Ramirez-Bajo MJ, Rovira J, Donor-derived cell-free DNA shows high sensitivity for the diagnosis of pancreas graft rejection in simultaneous pancreas-kidney transplantation: Transplantation, 2022; 106(8); 1690-97
26. Yoo A, Riedel A, Qian I, An initial analysis of the baseline levels of dd-cfDNA after pancreas transplantation: A prospective study from high-volume centers in the United States: Transplant Direct, 2023; 9(4); e1459
27. Klein JA, Gupta A, Budhiraja P, Cibrik D, Utility of allosure monitoring in simultaneous kidney and pancreas recipients [abstract]: Am J Transplant, 2019; 19(Suppl 3); 655-56
28. Ali N, Stewart Z, Miles J, 343.9: Longitudinal monitoring of pancreas and kidney transplant recipients using donor-derived cell-free DNA: Transplantation, 2022; 106(9S); S332
29. Gadi VK, Nelson JL, Boespflug ND, Soluble donor DNA concentrations in recipient serum correlate with pancreas-kidney rejection: Clin Chem, 2006; 52(3); 379-82
30. Richmond ME, Zangwill SD, Kindel SJ, Donor fraction cell-free DNA and rejection in adult and pediatric heart transplantation: J Heart Lung Transplant, 2020; 39(5); 454-63
31. Beck J, Oellerich M, Schulz U, Donor-derived cell-free DNA is a novel universal biomarker for allograft rejection in solid organ transplantation: Transplant Proc, 2015; 47(8); 2400-3
32. Deng MC, Eisen HJ, Mehra MR, Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling: Am J Transplant, 2006; 6(1); 150-60
33. Kim PJ, Olymbios M, Siu A, A novel donor-derived cell-free DNA assay for the detection of acute rejection in heart transplantation: J Heart Lung Transplant, 2022; 41(7); 919-27
34. Amadio JM, Rodenas-Alesina E, Superina S, Sparing the prod: Providing an alternative to endomyocardial biopsies with noninvasive surveillance after heart transplantation during COVID-19: CJC Open, 2022; 4(5); 479-87
35. Rodgers N, Gerding B, Cusi V, Comparison of two donor-derived cell-free DNA tests and a blood gene-expression profile test in heart transplantation: Clin Transplant, 2023; 37(6); e14984
36. Henricksen EJ, Moayedi Y, Purewal S, Combining donor derived cell free DNA and gene expression profiling for non-invasive surveillance after heart transplantation: Clin Transplant, 2023; 37(3); e14699
37. Agbor-Enoh S, Wang Y, Tunc I, Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation: EBioMedicine, 2019; 40; 541-53
38. De Vlaminck I, Valantine HA, Snyder TM, Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection: Sci Transl Med, 2014; 6(241); 241ra77
39. Magnusson JM, Ricksten A, Dellgren G, Cell-free DNA as a biomarker after lung transplantation: A proof-of-concept study: Immun Inflamm Dis, 2022; 10(5); e620
40. Bloom RD, Bromberg JS, Poggio ED, Cell-free DNA and active rejection in kidney allografts: J Am Soc Nephrol, 2017; 28(7); 2221-32
41. Hinojosa RJ, Chaffin K, Gillespie M, Villarreal VH, Donor-derived cell-free DNA may confirm real-time response to treatment of acute rejection in renal transplant recipients: Transplantation, 2019; 103(4); e61
42. Huang E, Gillespie M, Ammerman N, Donor-derived cell-free DNA combined with histology improves prediction of estimated glomerular filtration rate over time in kidney transplant recipients compared with histology alone: Transplant Direct, 2020; 6(8); e580
43. Martuszewski A, Paluszkiewicz P, Krol M, Donor-derived cell-free DNA in kidney transplantation as a potential rejection biomarker: A systematic literature review: J Clin Med, 2021; 10(2); 193
44. Stites E, Kumar D, Olaitan O, High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury: Am J Transplant, 2020; 20(9); 2491-98
45. Agbor-Enoh S, Shah P, Tunc I, Cell-free DNA to detect heart allograft acute rejection: Circulation, 2021; 143(12); 1184-97
46. Bu L, Gupta G, Pai A, Clinical outcomes from the assessing donor-derived cell-free DNA monitoring insights of kidney allografts with longitudinal surveillance (ADMIRAL) study: Kidney Int, 2022; 101(4); 793-803
47. Gray JN, Wolf-Doty T, Sulejmani N, KidneyCare guided immuno-optimization in renal allografts: The KIRA protocol: Methods Protoc, 2020; 3(4); 68
48. Dandamudi R, Gu H, Goss CW, Longitudinal evaluation of donor-derived cellfree DNA in pediatric kidney transplantation: Clin J Am Soc Nephrol, 2022; 17(11); 1646-55
49. Halloran PF, Reeve J, Madill-Thomsen KS, Combining donor-derived cell-free DNA fraction and quantity to detect kidney transplant rejection using molecular diagnoses and histology as confirmation: Transplantation, 2022; 106(12); 2435-42
50. Tedesco H, Sarwal R, Nakamura M, QSant, a multi-analyte urine-based test, types the injury of DGF and its recovery: Transplantation, 2022; 106(9S); S49
51. Allam S, Chuang P, Cooper M, Acute tubular injury and necrosis do not lead to meaningful elevations in donor-derived cell-free DNA (dd-cfDNA): Transplantation, 2022; 106(9S); S49-50
52. Weir M, Mandelbrot D, Bromberg J, Enhanced histological yield and actionable findings when biopsy is guided by donor-derived cell-free DNA (dd-cfDNA) kidney transplant: Transplantation, 2022; 106(9); S50-51
53. Shihab F, Brennan D, Ellis M, Wide spectrum of molecular injury highlights heterogeneity of Banff tubulitis and interstitial inflammation lesions: Transplantation, 2022; 106(9S); S51
54. Obrişcă B, Butiu M, Sibulesky L, Combining donor-derived cell-free DNA and donor specific antibody testing as non-invasive biomarkers for rejection in kidney transplantation: Sci Rep, 2022; 12; 15061
55. Guo K, Zhao L, Kleiboeker S, Friedewald J, Dynamic kidney graft failure risk prediction using serial donor derived cell free DNA: Transplantation, 2022; 106(9S); S54
56. Ramirez-Bajo MJ, Rovira J, Banon-maneus E, 232.10: Dynamic of donor derived cell-free DNA and blood gene expression after pancreas transplantation: Transplantation, 2022; 106(9S); S84
57. Miles J, Tatapudi V, Mattoo A, Longitudinal surveillance of a kidney transplant recipient during pregnancy using quantification of fetal and donor-derived cell-free DNA: Transplantation, 2022; 106(9S); S85
58. Sedej I, Štalekar M, Žnidarič MT, Urinary extracellular vesicle DNA cargo reflects kidney allograft injury: Transplantation, 2022; 106(9S); S55
59. Lui YY, Woo KS, Wang AY, Origin of plasma cell-free DNA after solid organ transplantation: Clin Chem, 2003; 49(3); 495-96
60. Macher HC, Suarez-Artacho G, Guerrero JM, Monitoring of transplanted liver health by quantification of organ-specific genomic marker in circulating DNA from receptor: PLoS One, 2014; 9(12); e113987
61. De Vlaminck I, Martin L, Kertesz M, Noninvasive monitoring of infection and rejection after lung transplantation: Proc Natl Acad Sci USA, 2015; 112(43); 13336-41
62. Bloom RD, Bromberg JS, Poggio ED, Cell-free DNA and active rejection in kidney allografts: J Am Soc Nephrol, 2017; 28(7); 2221-32
63. Ragalie WS, Stamm K, Mahnke D, Noninvasive assay for donor fraction of cell-free DNA in pediatric heart transplant recipients: J Am Coll Cardiol, 2018; 71(25); 2982-83
64. Whitlam JB, Ling L, Skene A, Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction: Am J Transplant, 2019; 19(4); 1037-49
65. Huang E, Sethi S, Peng A, Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients: Am J Transplant, 2019; 19(6); 1663-70
66. Khush KK, Patel J, Pinney S, Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study: Am J Transplant, 2019; 19(10); 2889-99
67. North PE, Ziegler E, Mahnke DK, Cell-free DNA donor fraction analysis in pediatric and adult heart transplant patients by multiplexed allele-specific quantitative PCR: Validation of a rapid and highly sensitive clinical test for stratification of rejection probability: PLoS One, 2020; 15(1); e0227385
68. Dauber EM, Kollmann D, Kozakowski N, Quantitative PCR of INDELs to measure donor-derived cell-free DNA-a potential method to detect acute rejection in kidney transplantation: A pilot study: Transpl Int, 2020; 33(3); 298-309
69. Gielis EM, Ledeganck KJ, Dendooven A, The use of plasma donor-derived, cell-free DNA to monitor acute rejection after kidney transplantation: Nephrol Dial Transplant, 2020; 35(4); 714-21
70. Zhang H, Zheng C, Li X, Diagnostic performance of donor-derived plasma cell-free DNA fraction for antibody-mediated rejection in post-renal transplant recipients: A prospective observational study: Front Immunol, 2020; 11; 342
71. Puliyanda DP, Swinford R, Pizzo H, Donor-derived cell-free DNA (dd-cfDNA) for detection of allograft rejection in pediatric kidney transplants: Pediatr Transplant, 2021; 25(2); e13850
72. Sayah D, Weigt SS, Ramsey A, Plasma donor-derived cell-free DNA levels are increased during acute cellular rejection after lung transplant: pilot data: Transplant Direct, 2020; 6(10); e608
73. Zhou Q, Liu F, Guo L, A novel urine cell-free DNA preservation solution and its application in kidney transplantation: Nephrology, 2021; 26(8); 684-91
74. Khush KK, De Vlaminck I, Luikart H, Donor-derived, cell-free DNA levels by next-generation targeted sequencing are elevated in allograft rejection after lung transplantation: ERJ Open Res, 2021; 7(1); 00462-2020
75. Jang MK, Tunc I, Berry GJ, Donor-derived cell-free DNA accurately detects acute rejection in lung transplant patients, a multicenter cohort study: J Heart Lung Transplant, 2021; 40(8); 82283
76. Kueng N, Arcioni S, Sandberg F, Comparison of methods for donor-derived cell-free DNA quantification in plasma and urine from solid organ transplant recipients: Front Genet, 2023; 14; 1089830
77. Chen XT, Qiu J, Wu ZX, Using both plasma and urine donor-derived cell-free DNA to identify various renal allograft injuries: Clin Chem, 2022; 68(6); 814-25
78. Edwards RL, Menteer J, Lestz RM, Baxter-Lowe LA, Cell-free DNA as a solid-organ transplant biomarker: Technologies and approaches: Biomark Med, 2022; 16(5); 401-15
Figures
Tables
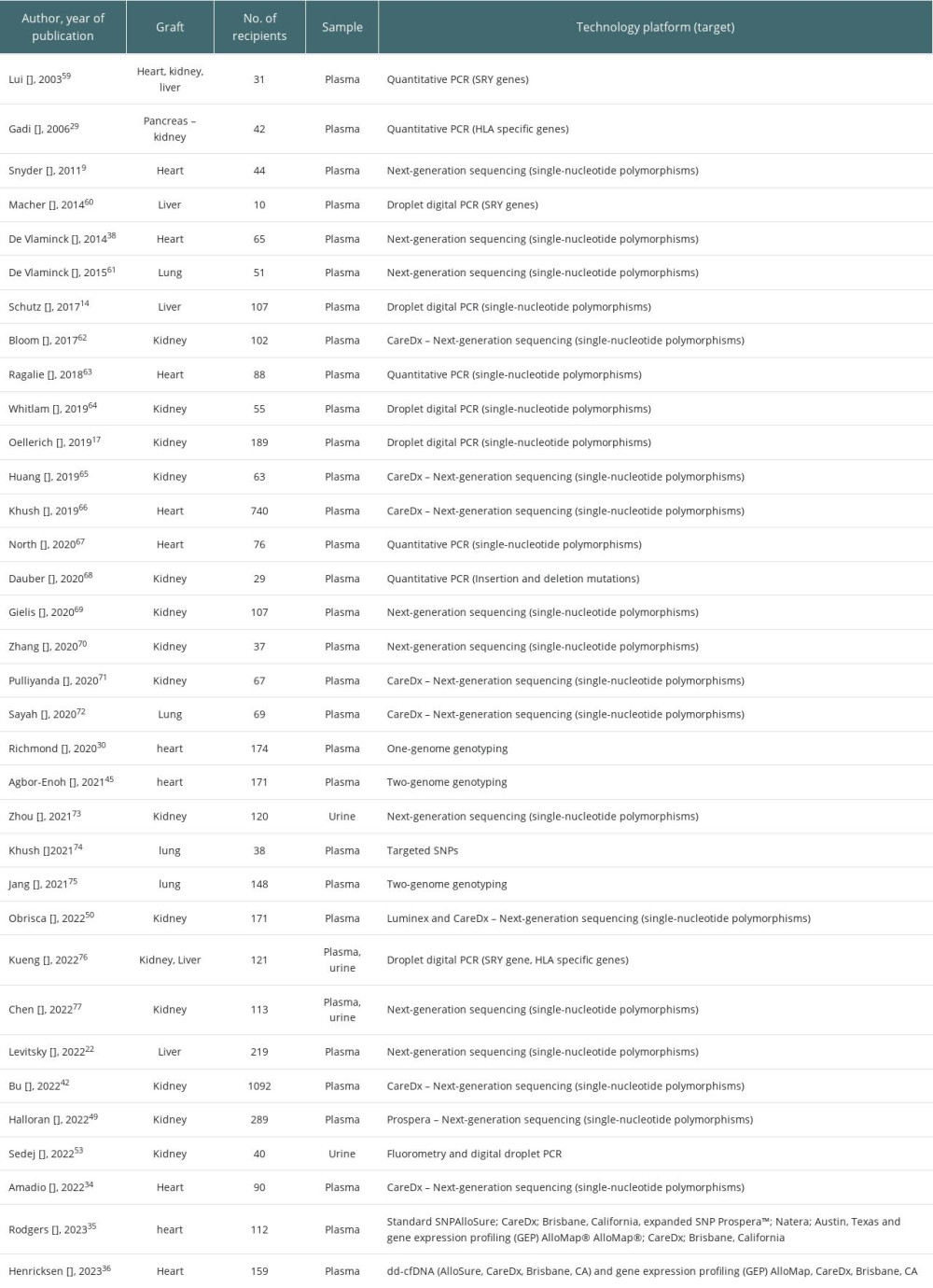 Table 1. Summary of the studies on cfDNA in regard to type of organ, source (plasma/urine or both) and technology platform used.
Table 1. Summary of the studies on cfDNA in regard to type of organ, source (plasma/urine or both) and technology platform used.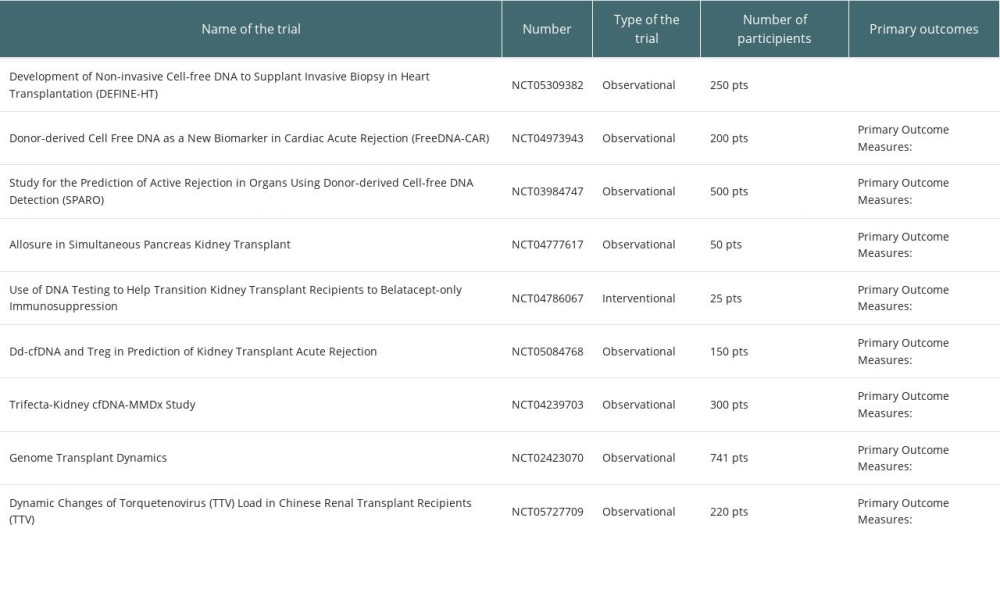 Table 2. Clinical trials with cfDNA in solid organ transplantation actually recruting patients.
Table 2. Clinical trials with cfDNA in solid organ transplantation actually recruting patients. Table 1. Summary of the studies on cfDNA in regard to type of organ, source (plasma/urine or both) and technology platform used.
Table 1. Summary of the studies on cfDNA in regard to type of organ, source (plasma/urine or both) and technology platform used. Table 2. Clinical trials with cfDNA in solid organ transplantation actually recruting patients.
Table 2. Clinical trials with cfDNA in solid organ transplantation actually recruting patients. In Press
28 Mar 2024 : Original article
Association Between FEV₁ Decline Rate and Mortality in Long-Term Follow-Up of a 21-Patient Pilot Clinical T...Ann Transplant In Press; DOI: 10.12659/AOT.942823
02 Apr 2024 : Original article
Liver Transplantation from Brain-Dead Donors with Hepatitis B or C in South Korea: A 2014-2020 Korean Organ...Ann Transplant In Press; DOI: 10.12659/AOT.943588
02 Apr 2024 : Original article
Effect of Dexmedetomidine Combined with Remifentanil on Emergence Agitation During Awakening from Sevoflura...Ann Transplant In Press; DOI: 10.12659/AOT.943281
03 Apr 2024 : Review article
Alternative Therapies in Transplantology as a Promising Perspective in MedicineAnn Transplant In Press; DOI: 10.12659/AOT.943387
Most Viewed Current Articles
05 Apr 2022 : Original article
Impact of Statins on Hepatocellular Carcinoma Recurrence After Living-Donor Liver TransplantationDOI :10.12659/AOT.935604
Ann Transplant 2022; 27:e935604
12 Jan 2022 : Original article
Risk Factors for Developing BK Virus-Associated Nephropathy: A Single-Center Retrospective Cohort Study of ...DOI :10.12659/AOT.934738
Ann Transplant 2022; 27:e934738
22 Nov 2022 : Original article
Long-Term Effects of Everolimus-Facilitated Tacrolimus Reduction in Living-Donor Liver Transplant Recipient...DOI :10.12659/AOT.937988
Ann Transplant 2022; 27:e937988
15 Mar 2022 : Case report
Combined Liver, Pancreas-Duodenum, and Kidney Transplantation for Patients with Hepatitis B Cirrhosis, Urem...DOI :10.12659/AOT.935860
Ann Transplant 2022; 27:e935860