26 July 2022: Original Paper
Donor CYP3A5 Expression Decreases Renal Transplantation Outcomes in White Renal Transplant Recipients
Karola Warzyszyńska1ABCDEFG*, Michał ZawistowskiDOI: 10.12659/AOT.936276
Ann Transplant 2022; 27:e936276
Abstract
BACKGROUND: After renal transplantation, immunosuppressants should be administered to prevent organ rejection and prolong graft survival. One of them is tacrolimus, which is metabolized by the CYP3A enzyme family. The variability of the CYP3A5 gene in renal transplant recipients has been previously studied for its correlation with acute rejection and allogeneic kidney function. CYP3A5 enzyme is also present in the renal tissue, and its relevance has not yet been extensively investigated. This study aimed to evaluate the effect of donor and recipient CYP3A5 expression status on early and long-term transplant outcomes.
MATERIAL AND METHODS: Single-nucleotide polymorphism in CYP3A5 (rs776746) was analyzed in 95 kidney transplant recipients and their grafts. The effect of donor and recipient genotypes on the primary endpoint, which was the loss of the renal graft over 5-year follow-up, was assessed. The secondary endpoints were biopsy-proven acute rejection, proteinuria, delayed graft function, and renal function.
RESULTS: Patients who received a CYP3A5*1 allele-carrying kidney (n=16) were at greater risk of graft loss (adjusted hazard ratio, 95% CI: 10.61, 2.28-49.42, P=.003) than those with the CYP3A5*3/*3 genotype (n=79). Renal CYP3A5 expression was also a predictor of acute rejection between the 2nd and 12th post-transplant months (adjusted odds ratio, 95% CI: 4.36; 1.08-17.6, P=.038) and proteinuria at different time intervals. No effect of the recipient CYP3A5 genotype was observed.
CONCLUSIONS: The donor CYP3A5 genotype is associated with inferior transplantation outcomes. Local renal tacrolimus metabolism is a potential target for improving long-term transplantation outcomes.
Keywords: CYP3A5 Protein, Human, Delayed Graft Function, Graft Rejection, Graft Survival, Kidney Transplantation, Proteinuria, Cytochrome P-450 CYP3A, Genotype, Humans, Immunosuppressive Agents, Kidney, Polymorphism, Single Nucleotide, Tacrolimus, transplant recipients
Background
Calcineurin inhibitors are among the most important groups of immunosuppressants, with tacrolimus (TAC) being the first-choice agent for the treatment of renal transplant recipients (RTRs) [1]. When TAC was placed on the market, it reduced biopsy-proven acute rejection (BPAR) occurrence at 12-month follow-up to 10–20%. Early BPAR is a crucial factor affecting overall patient and allogeneic graft survival [2,3].
TAC dosage adjustment remains a challenge owing to its narrow therapeutic index and high variability in pharmacokinetics. Underexposure favors allograft rejection, whereas overexposure results in severe adverse effects. Treatment requires therapeutic drug monitoring using whole-blood trough TAC concentration (C0) measures [1,4].
It is primarily metabolized by CYP3A cytochromes localized in the liver and gut mucosa. The function of CYP3A5 is well documented. The expression of CYP3A5 enzymes, which accelerate TAC metabolism and elimination, depends on the
Therefore, it was expected that the recipient’s
Our cohort study aimed to assess whether the
Material and Methods
STUDY COHORT:
We conducted a retrospective cohort study in 95 deceased donor kidney recipients who were selected from among 595 patients who underwent transplantation between January 2010 and January 2017 at a single center. Patients were considered eligible for inclusion based on the following criteria: 1) >18 years old; 2) donation after brainstem death donor; 3) White ethnicity of both donor and recipient; 4) receiving twice-daily TAC formula (Prograf®, Astellas Pharma, Warsaw, Poland) in post-transplant immunosuppressive treatment; 5) availability of donor DNA; 6) completeness of the follow-up (patients who were managed at our outpatient transplantation clinic); and 7) written informed consent to participate in the study. Exclusion criteria were: 1) multiorgan transplantation and 2) primary nonfunctioning kidney transplant. The participants, transplanted 1–7 years prior to recruitment and with stable transplant function, were consecutively enrolled during their follow-up visits at our outpatient transplantation clinic between December 2018 and March 2019 until the required sample size was reached. Recipient blood samples were collected during routine check-up visits after obtaining informed consent. Donor deoxynucleic acid (DNA) was obtained from the tissue typing laboratory of the Department of Immunology, where samples were isolated and stored at −20°C after routine crossmatching before transplantation. Demographic data of the RTRs and their corresponding donors, drug doses, TAC C0 results, and clinical outcomes of transplantation were obtained from medical records. Donors were evaluated using the non-scaled US Kidney Donor Risk Index (KDRI) [22]. No deaths were noted in the observed population, and no patients were lost to follow-up.
The analyzed subjects received triple immunosuppressive therapy consisting of:
In those patients who had positive panel reactive antibody test (PRA) or underwent re-transplantation, the induction therapy was implemented: either anti-thymocyte globulin (Thymoglobulin) or basiliximab. The immunosuppressive regimen was conducted in line with the Polish Transplantation Society guidelines [23].
Whole-blood C0 TAC concentrations were assayed using the chemiluminescence microparticle method and measured routinely during the post-transplant check-ups. This study conforms to the STREGA statement [24].
GENOTYPING:
Real-time polymerase chain reaction (RT-PCR) was used to determine the genotype of CYP3A5 (rs776746; g.99672916C>T) single-nucleotide polymorphism (SNP) in deceased donors and RTRs. RTR DNA was extracted from whole blood using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany). The quality and quantity of the DNA samples were verified by electrophoresis using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Analysis was performed using the rs776746 TaqMan® SNP Genotyping Assay (Thermo Fisher Scientific, Vilnius, Lithuania), according to the qPCR protocol for SNP Genotyping (Thermo Fisher Scientific, Carlsbad, US) [25]. RT-PCR was performed using the QuantStudio™ 12 K Flex Real-Time PCR System (Thermo Fisher Scientific, Carlsbad, US) and repeated 3 times to minimize any genotyping errors. Data analysis was performed using TaqMan® Genotyper software (Thermo Fisher Scientific).
OUTCOME MEASURES:
We compared subgroups defined by (1) donor and (2) recipient CYP3A5 expressor status. The primary outcome was graft loss observed within 5 post-transplant years. It was defined as a loss of renal function resulting in the need for long-term dialysis, graftectomy, or re-transplantation [26]. Secondary outcomes included:
ETHICS:
The study protocol was reviewed and approved by the institutional review board of the Medical University of Warsaw (IRB protocol number: KB/203/2018). This study conforms to the Declaration of Helsinki, Council for International Organizations of Medical Sciences Guidelines, and International Conference on Harmonization of Good Clinical Practice.
STATISTICAL ANALYSIS:
Continuous variables were tested for normality using the Shapiro-Wilk test along with the Q-Q plot analysis and reported as mean (M) and standard deviation (SD) or median (Mdn) with the first and third quartile (interquartile range, IQR), as appropriate. Categorical data were reported as the number of events and percentages. Hardy-Weinberg equilibrium and the chi-squared test were used to evaluate the allele frequencies. Levene’s test was used to assess the equality of variances. Differences between subgroups were tested using the t test, Welch’s t test, or Mann-Whitney U test for continuous variables and the chi-squared or Fisher’s exact test for categorical variables. The primary outcome was evaluated using Kaplan-Meier curves, log-rank test, and Cox proportional hazards analysis. Logistic regression was used to estimate secondary endpoints. All multivariable models were created using backward elimination. Potential risk factors were selected based on a literature search [2,3,28,31,32]. Owing to the small sample size, we could only include a limited number of covariates in our models. Therefore, in some cases, 2 alternative models were created. Missing data were deleted pairwise. Statistical significance was defined as a 2-sided P value <.05. All tests and plots were created using the survival, survminer, transplantr, and CoxR2 packages in R 4.1.2 (R Core Team, 2021).
Results
GENOTYPING, DONOR, AND PATIENT CHARACTERISTICS:
The 95 recipients received kidneys from 63 donors. The call rate of the tested samples from both donors and recipients was 100% (n=158). CYP3A5 *1/*3 (TC) was identified in 22 (13.9%) and *3/*3 (CC) in 136 (86.1%) individuals. No wild-type homozygotes *1/*1 (TT) were found in the tested samples. The genotype distribution was consistent with the HWE χ2 (2, 158)=0.9, P=.642. The allele frequencies were T=0.070 and C=0.930, which is consistent with the data from the 1000 Genomes Project [5] for the European population (T=0.057 and C=0.943). The CYP3A5 genotypes were divided into 2 subgroups: expressors (CYP3A5 *1/*3) and non-expressors (CYP3A5 *3/*3). There were 8 (12.7%) and 14 (14.7%) CYP3A5 expressors among donors and recipients, respectively.
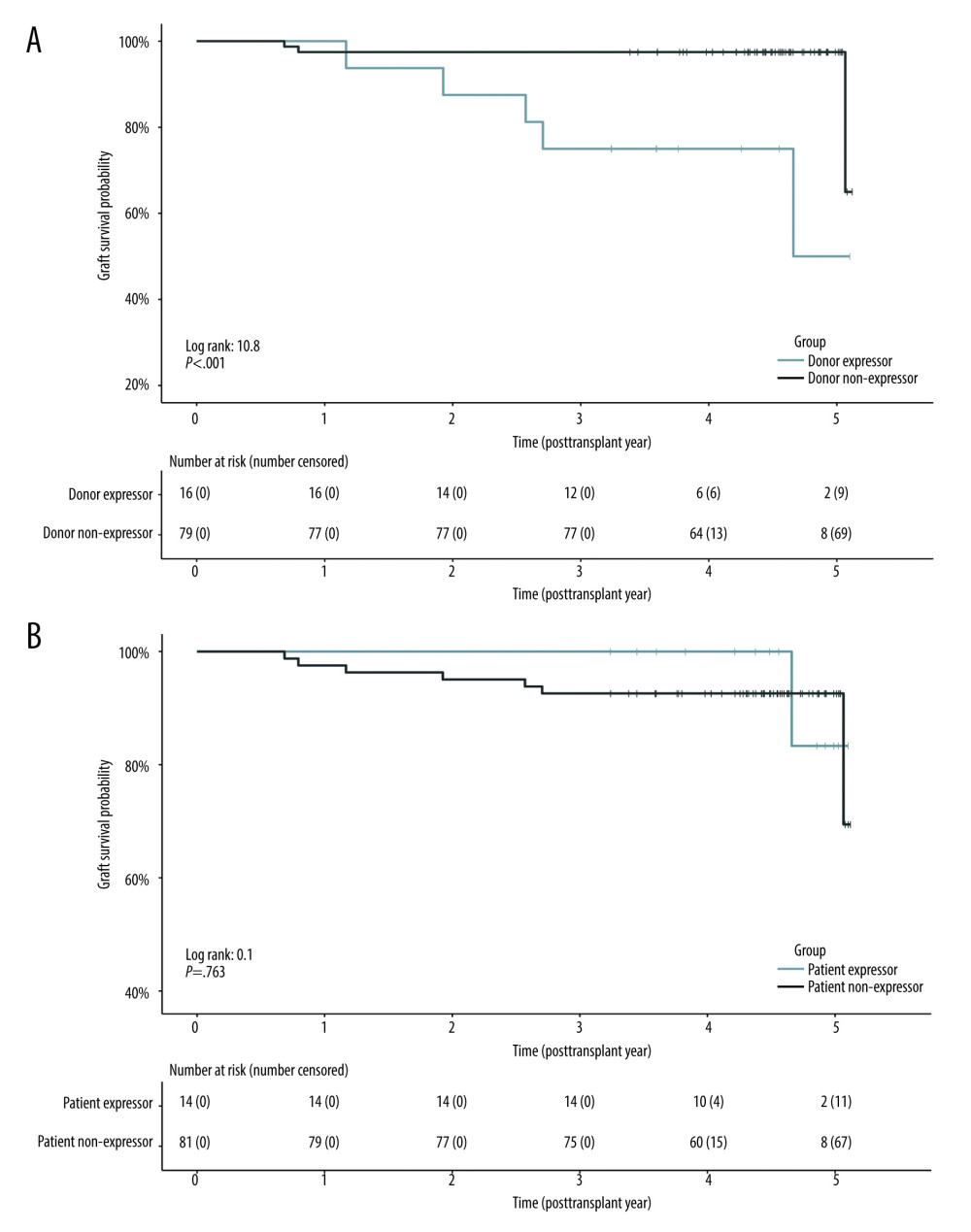
RTRs were divided into subgroups according to the presence or absence of CYP3A5 coding alleles in 1) donors (D+, donor expressor; D−, donor non-expressor) and 2) recipients (R+, recipient expressor; R−, recipient non-expressor). The demographic and clinical characteristics are provided in Table 1.
ASSOCIATION OF POLYMORPHISMS WITH TAC EXPOSURE:
The median TAC C0 levels according to CYP3A5 expression in both recipients and donors within 3 years of follow-up are shown in Supplementary Figure 1.
In recipients who received kidneys from CYP3A5 expressors, TAC C0 was significantly lower on the third day after transplantation than in non-expressors (Mdn [IQR], 6.0 [4.5–6.7] ng/mL vs 10.9 [6.8–16.5] ng/mL, respectively; U = 248.50,
ASSOCIATION BETWEEN POLYMORPHISMS AND TRANSPLANTATION OUTCOMES:
The parameters considered relevant for transplantation outcomes, such as graft loss, BPAR, proteinuria, and DGF, were assessed in CYP3A5 expressing grafts and recipients, and are summarized in Table 2 and Supplementary Figure 2.
GRAFT LOSS: We conducted survival analysis to evaluate the occurrence of graft loss in predefined subgroups (Figure 1). The number of events was 8 (8.4%) within the 5-year follow-up. We found that RTRs with grafts from CYP3A5 expressors were at greater risk of graft loss (Figure 1A), log-rank test, χ2 (1)=10.8, P<.001. Cox proportional hazard analysis revealed donor CYP3A5 expressor status to be a risk factor for graft loss when adjusted for unscaled US KDRI (adjusted hazard ratio, 95% confidence interval [CI]; 10.61 (2.28–49.42), P=.003, Table 3). We also created a Cox model adjusted for BPAR episodes, which also significantly increased the risk of loss of graft function; however, the model was overfitted due to the small number of observations (Supplementary Table 1).
SECONDARY OUTCOMES:
BPAR episodes occurred in 12 patients (12.6%) within the first postoperative year. Logistic regression analysis showed that donor
Donor CYP3A5 expressor status also increased the risk of proteinuria within the first post-transplant year (adjusted OR, 95%CI: 5.90, 1.40–24.82; P=.015, Table 4) and after that time, within the study follow-up (adjusted OR, 95%CI; 4.49, 1.28–15.72; P=.019). No differences were detected in proteinuria within the first post-transplant month and in delayed graft function. Owing to the small sample size and number of events, analysis of other endpoints was unfeasible. No significant associations were detected regarding recipient CYP3A5 expressor status.
The function of the allogeneic kidney was evaluated using eGFR during 3 years of follow-up. No significant difference was observed, regardless of donor CYP3A5 expression, until the third post-transplant year, when recipient CYP3A5 expression was associated with lower eGFR (M [SD], 59.55 [21.01] ml/min/1.73 m2 vs 45.46 [16.73] ml/min/1.73 m2; t[87]=−2.37, P=.020) (Supplementary Table 2).
Discussion
LIMITATIONS:
This study has several limitations, among which the most important are its retrospective design and the small number of patients included in the analyses. As the funding was limited, we were only able to include a limited number of patients, which might have the same impact, mainly on the secondary analyses. Moreover, in our center, no protocol biopsies are routinely performed, resulting in possible underestimation of BPAR occurrence. We are aware that the method of choice in TAC C0 determination is LC-MS/MS, and the use of immunoenzymatic assay may provide a measurement error, but it was not available in our center when the data were collected. Finally, the results of our study cannot be extrapolated to other populations owing to the different prevalence of the evaluated polymorphisms worldwide.
Conclusions
In conclusion, our data showed that the donor
Tables
Table 1. Baseline demographics and clinicopathological characteristics of patients and brain-dead kidney donors.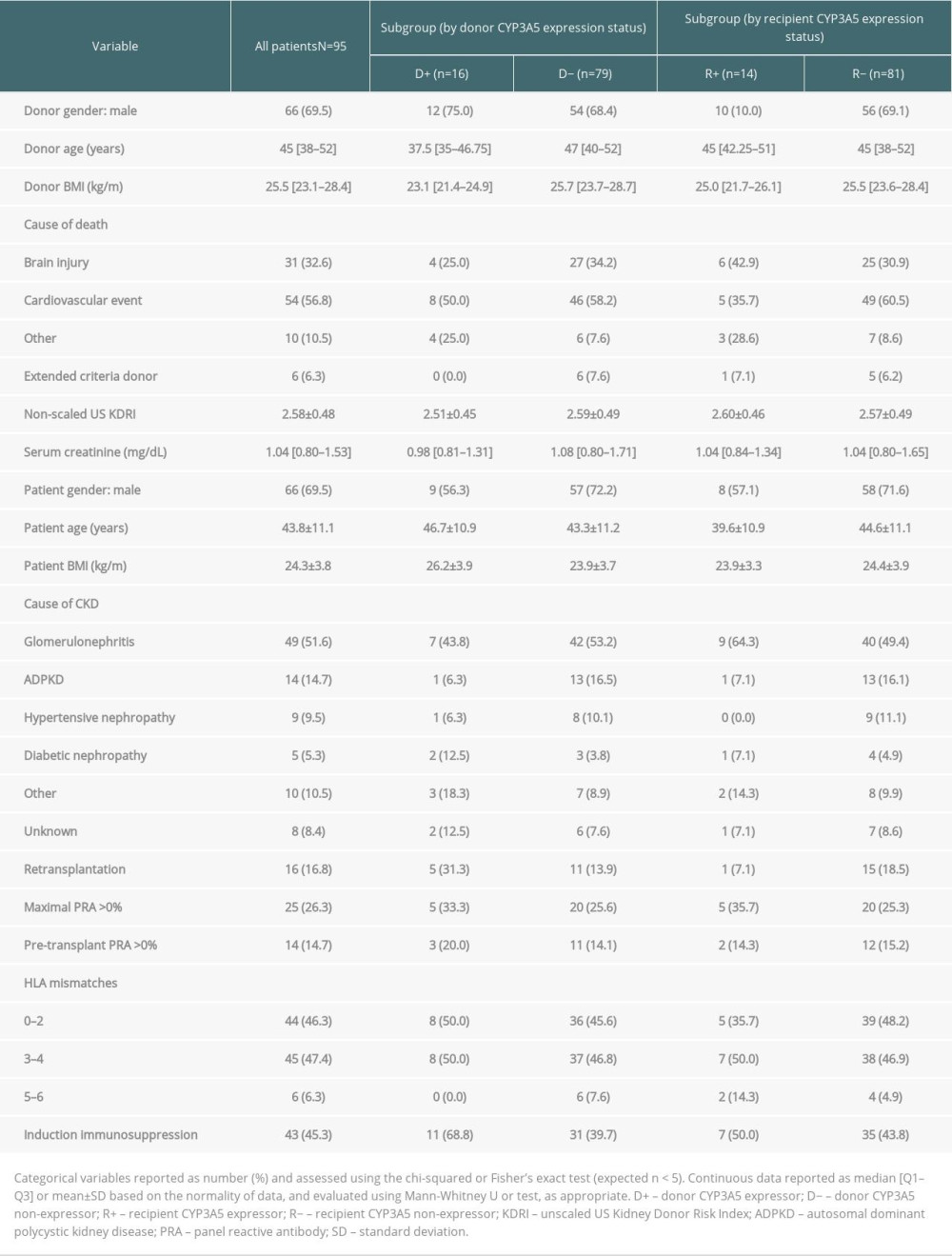 Table 2. Transplantation outcomes and group comparisons.
Table 2. Transplantation outcomes and group comparisons.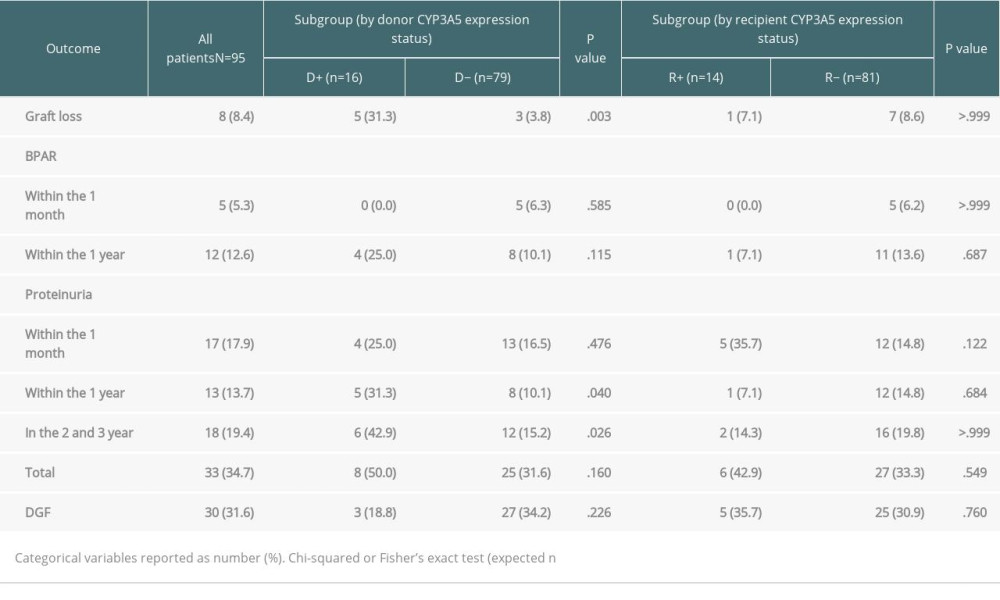 Table 3. Cox proportional hazards regression analysis for graft loss.
Table 3. Cox proportional hazards regression analysis for graft loss.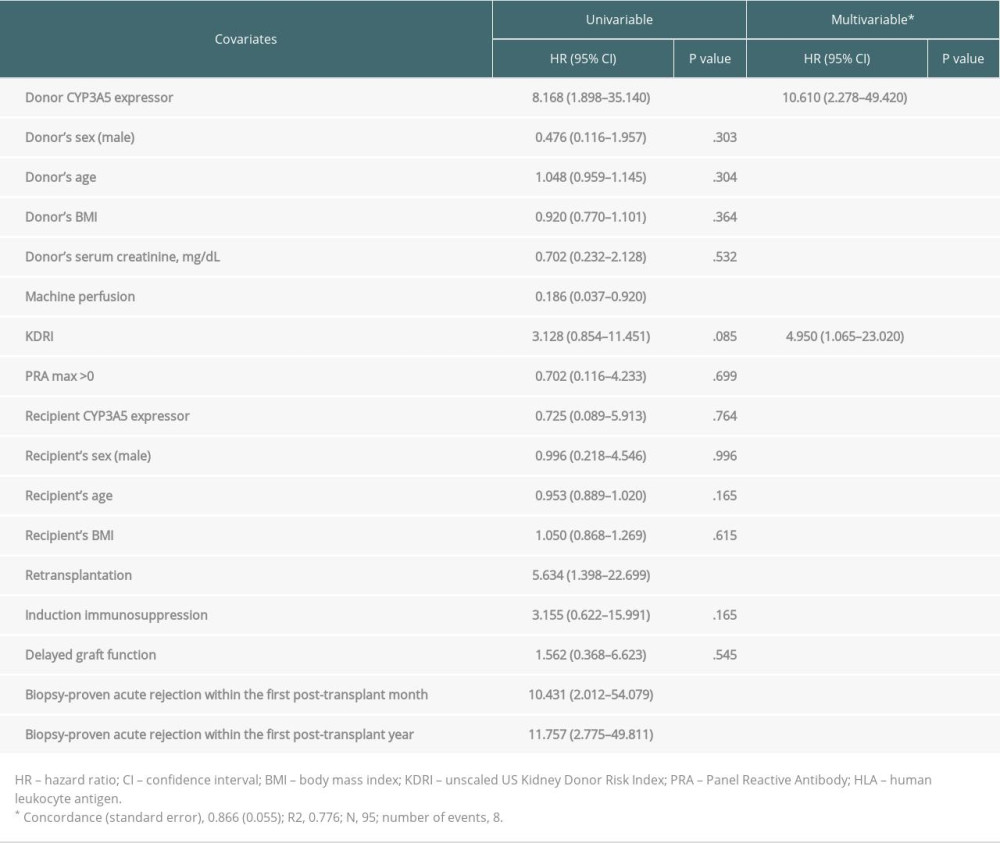 Table 4. Logistic regression models to estimate secondary endpoints (biopsy-proven acute rejection, proteinuria, and delayed graft function) using donor CYP3A5 expressor status as a predictive factor.
Table 4. Logistic regression models to estimate secondary endpoints (biopsy-proven acute rejection, proteinuria, and delayed graft function) using donor CYP3A5 expressor status as a predictive factor.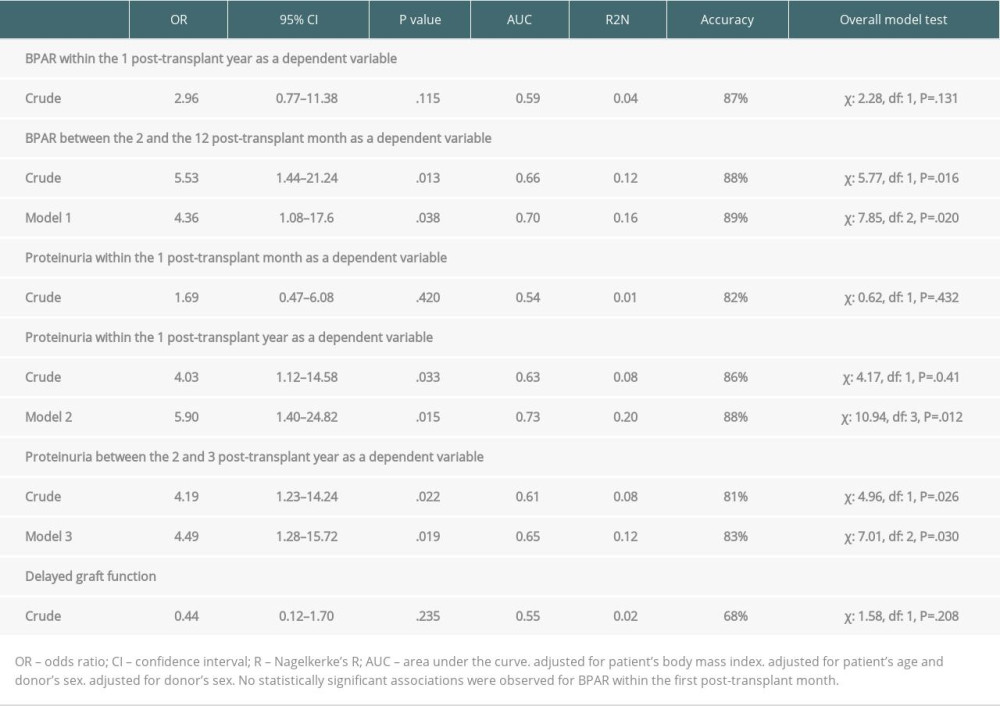 Supplementary Table 1. Cox proportional hazards regression analysis for graft loss adjusted for biopsy-proven acute rejection episodes.
Supplementary Table 1. Cox proportional hazards regression analysis for graft loss adjusted for biopsy-proven acute rejection episodes.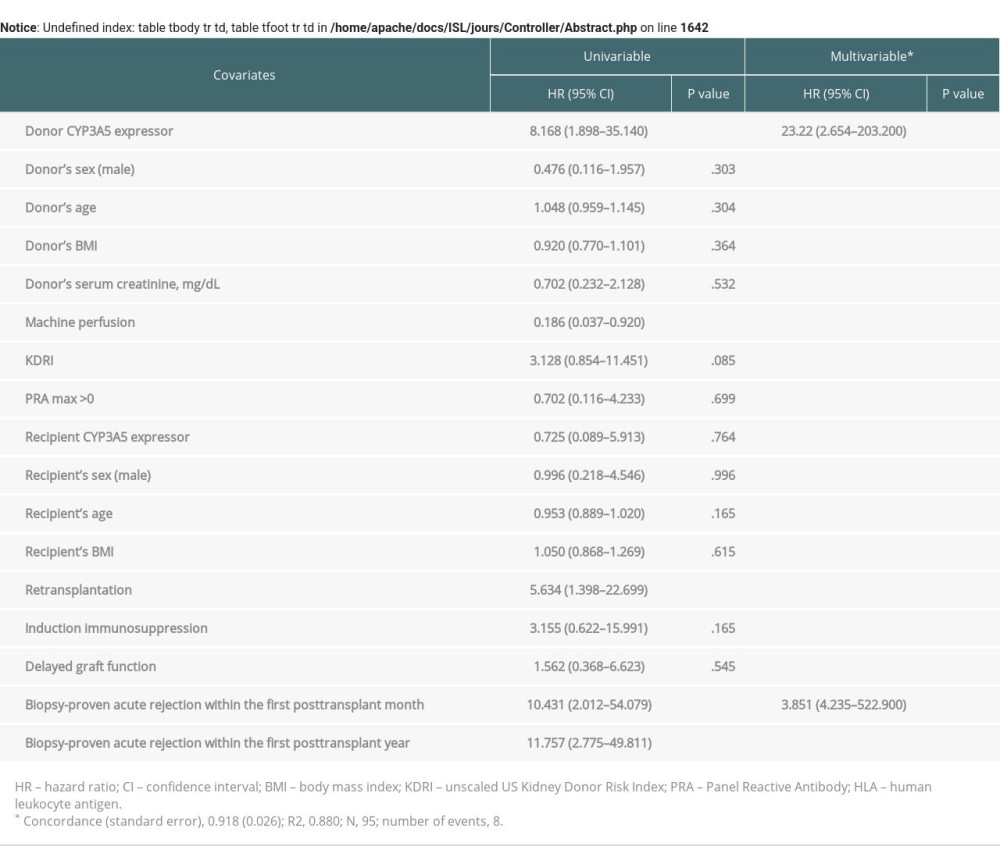 Supplementary Table 2. Estimated glomerular filtration rate (eGFR) within three-year follow-up depending on the presence of the renal or recipient CYP3A5*1 allele variant.
Supplementary Table 2. Estimated glomerular filtration rate (eGFR) within three-year follow-up depending on the presence of the renal or recipient CYP3A5*1 allele variant.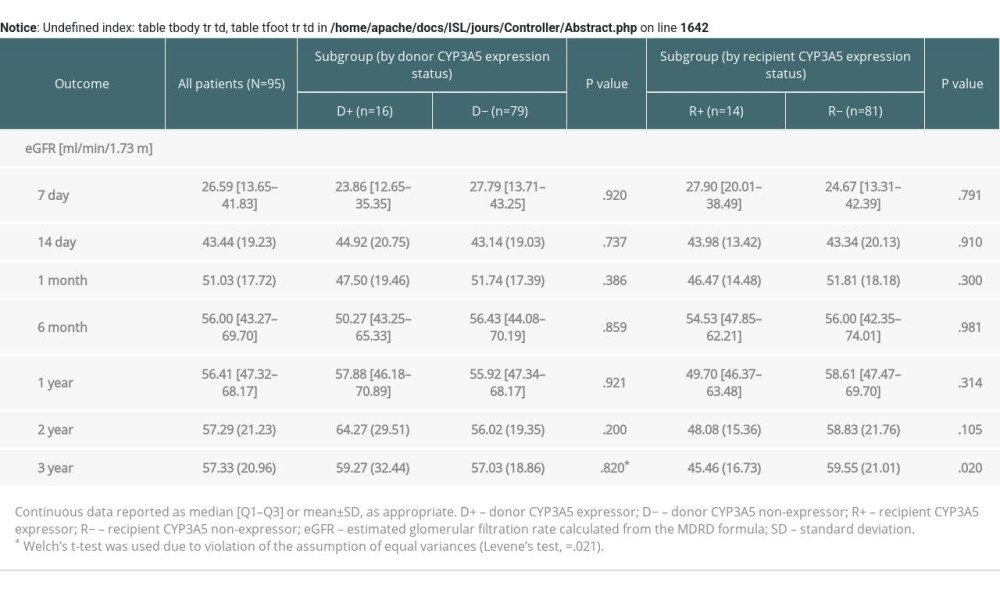
References
1. Baker RJ, Mark PB, Patel RK, Renal association clinical practice guideline in post-operative care in the kidney transplant recipient: BMC Nephrol, 2017; 18(1); 174
2. Clayton PA, McDonald SP, Russ GR, Chadban SJ, Long-term outcomes after acute rejection in kidney transplant recipients: An ANZDATA analysis: J Am Soc Nephrol, 2019; 30(9); 1697-707
3. Lemoine M, Titeca Beauport D, Lobbedez T, Risk factors for early graft failure and death after kidney transplantation in recipients older than 70 years: Kidney Int Rep, 2019; 4(5); 656-66
4. Brunet M, van Gelder T, Asberg A, Therapeutic drug monitoring of tacrolimus-personalized therapy: Second consensus report: Ther Drug Monit, 2019; 41(3); 261-307
5. Auton A, Brooks LS, Durbin RM1000 Genomes Project Consortium, A global reference for human genetic variation: Nature, 2015; 526; 68-74
6. Rojas L, Neumann I, Herrero MJ, Effect of CYP3A5*3 on kidney transplant recipients treated with tacrolimus: A systematic review and meta-analysis of observational studies: Pharmacogenomics J, 2015; 15(1); 38-48
7. Zong YP, Wang ZJ, Zhou WL, Effects of CYP3A5 polymorphisms on tacrolimus pharmacokinetics in pediatric kidney transplantation: A systematic review and meta-analysis of observational studies: World J Pediatr, 2017; 13(5); 421-26
8. Khan AR, Raza A, Firasat S, Abid A, CYP3A5 gene polymorphisms and their impact on dosage and trough concentration of tacrolimus among kidney transplant patients: A systematic review and meta-analysis: Pharmacogenomics J, 2020; 20(4); 553-62
9. Flahault A, Anglicheau D, Loriot MA, Clinical impact of the CYP3A5 6986A>G allelic variant on kidney transplantation outcomes: Pharmacogenomics, 2017; 18(2); 165-73
10. Gervasini G, Garcia-Pino G, Vergara E, CYP3A genotypes of donors but not those of the patients increase the risk of acute rejection in renal transplant recipients on calcineurin inhibitors: A pilot study: Eur J Clin Pharmacol, 2018; 74(1); 53-60
11. Glowacki F, Lionet A, Buob D, CYP3A5 and ABCB1 polymorphisms in donor and recipient: impact on Tacrolimus dose requirements and clinical outcome after renal transplantation: Nephrol Dial Transplant, 2011; 26(9); 3046-50
12. Bolbrinker J, Seeberg S, Schostak M, CYP3A5 genotype-phenotype analysis in the human kidney reveals a strong site-specific expression of CYP3A5 in the proximal tubule in carriers of the CYP3A5*1 allele: Drug Metab Dispos, 2012; 40(4); 639-41
13. Dai Y, Hebert MF, Isoherranen N, Effect of CYP3A5 polymorphism on tacrolimus metabolic clearance in vitro: Drug Metab Dispos, 2006; 34(5); 836-47
14. Joy MS, Hogan SL, Thompson BD, Cytochrome P450 3A5 expression in the kidneys of patients with calcineurin inhibitor nephrotoxicity: Nephrol Dial Transplant, 2007; 22(7); 1963-68
15. Metalidis C, Lerut E, Naesens M, Kuypers DR, Expression of CYP3A5 and P-glycoprotein in renal allografts with histological signs of calcineurin inhibitor nephrotoxicity: Transplantation, 2011; 91(10); 1098-102
16. Udomkarnjananun S, Townamchai N, Chariyavilaskul P, The cytochrome P450 3A5 non-expressor kidney allograft as a risk factor for calcineurin inhibitor nephrotoxicity: Am J Nephrol, 2018; 47(3); 182-90
17. Xia T, Zhu S, Wen Y, Risk factors for calcineurin inhibitor nephrotoxicity after renal transplantation: A systematic review and meta-analysis: Drug Des Devel Ther, 2018; 12; 417-28
18. Naesens M, Lerut E, de Jonge H, Donor age and renal P-glycoprotein expression associate with chronic histological damage in renal allografts: J Am Soc Nephrol, 2009; 20(11); 2468-80
19. Hu R, Barratt DT, Coller JK, Effect of tacrolimus dispositional genetics on acute rejection in the first 2 weeks and estimated glomerular filtration rate in the first 3 months following kidney transplantation: Pharmacogenet Genomics, 2019; 29(1); 9-17
20. Yang L, de Winter BC, van Schaik RH, CYP3A5 and ABCB1 polymorphisms in living donors do not impact clinical outcome after kidney transplantation: Pharmacogenomics, 2018; 19(11); 895-903
21. Woillard JB, Gatault P, Picard N, A donor and recipient candidate gene association study of allograft loss in renal transplant recipients receiving a tacrolimus-based regimen: Am J Transplant, 2018; 18(12); 2905-13
22. Rao PS, Schaubel DE, Guidinger MK, A comprehensive risk quantification score for deceased donor kidneys: The Kidney Donor Risk Index: Transplantation, 2009; 88(2); 231-36
23. Durlik MZ, PTT Clinical Practice Guidelines for immunosuppression in organ transplant recipients: Polish Transplant Society Guidelines, 2016
24. Little J, Higgins JP, Ioannidis JP, STrengthening the REporting of Genetic Association Studies (STREGA) – an extension of the STROBE statement: Genet Epidemiol, 2009; 33(7); 581-98
25. Malkki M, Petersdorf EW, Genotyping of single nucleotide polymorphisms by 5′ nuclease allelic discrimination: Methods Mol Biol, 2012; 882; 173-82
26. Taber DJ, Gebregziabher M, Payne EH, Overall graft loss versus death-censored graft loss: Unmasking the magnitude of racial disparities in outcomes among us kidney transplant recipients: Transplantation, 2017; 101(2); 402-10
27. Garces JC, Giusti S, Staffeld-Coit C, Antibody-mediated eejection: A review: Ochsner J, 2017; 17(1); 46-55
28. Amer H, Cosio FG, Significance and management of proteinuria in kidney transplant recipients: J Am Soc Nephrol, 2009; 20(12); 2490-92
29. Lim MA, Bloom RD, Medical therapies to reduce delayed graft function and improve long-term graft survival: Are we making progress?: Clin J Am Soc Nephrol, 2020; 15(1); 13-15
30. Levey AS, Coresh J, Greene T, Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate: Ann Intern Med, 2006; 145(4); 247-54
31. Foroutan F, Friesen EL, Clark KE, Risk factors for 1-year graft loss after kidney transplantation. Systematic Review and Meta-Analysis: Clin J Am Soc Nephrol, 2019; 14(11); 1642-50
32. Zimmerman D, House AA, Kim SJ, The risk of acute rejection following kidney transplant by 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D status: A prospective cohort study: Can J Kidney Health Dis, 2017; 4; 2054358117699822
33. Fernández-Fresnedo G, Plaza JJ, Sánchez-Plumed J, Proteinuria: A new marker of long-term graft and patient survival in kidney transplantation: Nephrol Dial Transplant, 2004; 19(Suppl 3); iii47-iii51
34. Rekers NV, Flaig TM, Mallat MJK, Donor genotype and intragraft expression of CYP3A5 reflect the response to steroid treatment during acute renal allograft rejection: Transplantation, 2017; 101(9); 2017-25
35. Naesens M, Kuypers DR, Sarwal M, Calcineurin inhibitor nephrotoxicity: Clin J Am Soc Nephrol, 2009; 4(2); 481-508
36. Tavira B, Gómez J, Díaz-Corte C, The donor ABCB1 (MDR-1) C3435T polymorphism is a determinant of the graft glomerular filtration rate among tacrolimus treated kidney transplanted patients: J Hum Genet, 2015; 60; 273-76
Tables
 Table 1. Baseline demographics and clinicopathological characteristics of patients and brain-dead kidney donors.
Table 1. Baseline demographics and clinicopathological characteristics of patients and brain-dead kidney donors. Table 2. Transplantation outcomes and group comparisons.
Table 2. Transplantation outcomes and group comparisons. Table 3. Cox proportional hazards regression analysis for graft loss.
Table 3. Cox proportional hazards regression analysis for graft loss. Table 4. Logistic regression models to estimate secondary endpoints (biopsy-proven acute rejection, proteinuria, and delayed graft function) using donor CYP3A5 expressor status as a predictive factor.
Table 4. Logistic regression models to estimate secondary endpoints (biopsy-proven acute rejection, proteinuria, and delayed graft function) using donor CYP3A5 expressor status as a predictive factor. Table 1. Baseline demographics and clinicopathological characteristics of patients and brain-dead kidney donors.
Table 1. Baseline demographics and clinicopathological characteristics of patients and brain-dead kidney donors. Table 2. Transplantation outcomes and group comparisons.
Table 2. Transplantation outcomes and group comparisons. Table 3. Cox proportional hazards regression analysis for graft loss.
Table 3. Cox proportional hazards regression analysis for graft loss. Table 4. Logistic regression models to estimate secondary endpoints (biopsy-proven acute rejection, proteinuria, and delayed graft function) using donor CYP3A5 expressor status as a predictive factor.
Table 4. Logistic regression models to estimate secondary endpoints (biopsy-proven acute rejection, proteinuria, and delayed graft function) using donor CYP3A5 expressor status as a predictive factor. Supplementary Table 1. Cox proportional hazards regression analysis for graft loss adjusted for biopsy-proven acute rejection episodes.
Supplementary Table 1. Cox proportional hazards regression analysis for graft loss adjusted for biopsy-proven acute rejection episodes. Supplementary Table 2. Estimated glomerular filtration rate (eGFR) within three-year follow-up depending on the presence of the renal or recipient CYP3A5*1 allele variant.
Supplementary Table 2. Estimated glomerular filtration rate (eGFR) within three-year follow-up depending on the presence of the renal or recipient CYP3A5*1 allele variant. In Press
18 Mar 2024 : Original article
Does Antibiotic Use Increase the Risk of Post-Transplantation Diabetes Mellitus? A Retrospective Study of R...Ann Transplant In Press; DOI: 10.12659/AOT.943282
20 Mar 2024 : Original article
Transplant Nephrectomy: A Comparative Study of Timing and Techniques in a Single InstitutionAnn Transplant In Press; DOI: 10.12659/AOT.942252
28 Mar 2024 : Original article
Association Between FEV₁ Decline Rate and Mortality in Long-Term Follow-Up of a 21-Patient Pilot Clinical T...Ann Transplant In Press; DOI: 10.12659/AOT.942823
02 Apr 2024 : Original article
Liver Transplantation from Brain-Dead Donors with Hepatitis B or C in South Korea: A 2014-2020 Korean Organ...Ann Transplant In Press; DOI: 10.12659/AOT.943588
Most Viewed Current Articles
05 Apr 2022 : Original article
Impact of Statins on Hepatocellular Carcinoma Recurrence After Living-Donor Liver TransplantationDOI :10.12659/AOT.935604
Ann Transplant 2022; 27:e935604
12 Jan 2022 : Original article
Risk Factors for Developing BK Virus-Associated Nephropathy: A Single-Center Retrospective Cohort Study of ...DOI :10.12659/AOT.934738
Ann Transplant 2022; 27:e934738
22 Nov 2022 : Original article
Long-Term Effects of Everolimus-Facilitated Tacrolimus Reduction in Living-Donor Liver Transplant Recipient...DOI :10.12659/AOT.937988
Ann Transplant 2022; 27:e937988
15 Mar 2022 : Case report
Combined Liver, Pancreas-Duodenum, and Kidney Transplantation for Patients with Hepatitis B Cirrhosis, Urem...DOI :10.12659/AOT.935860
Ann Transplant 2022; 27:e935860